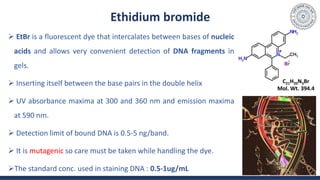
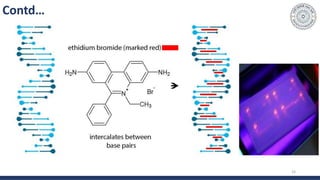
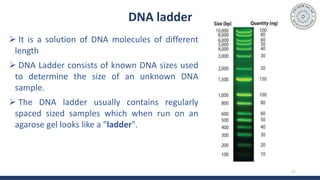
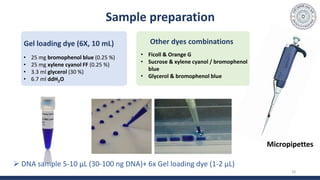
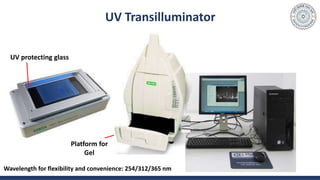
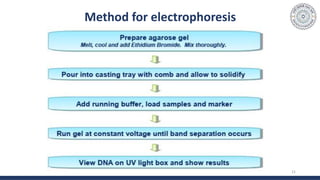
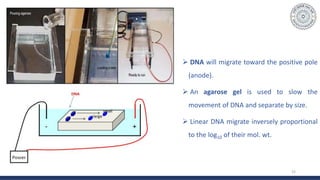
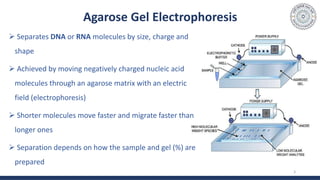
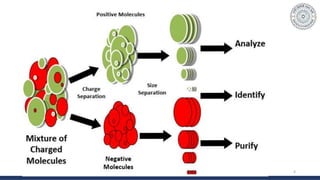
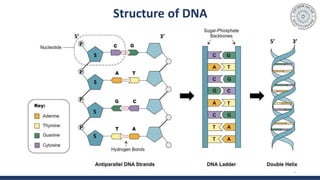
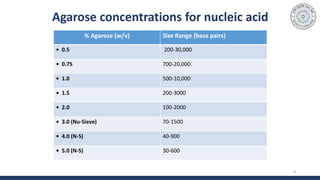
The document discusses agarose gel electrophoresis, which is used to separate DNA fragments by size. DNA samples are loaded onto an agarose gel and an electric current is applied, causing the negatively charged DNA to migrate through the gel at rates depending on fragment size. Smaller fragments move faster and travel farther than larger fragments. After electrophoresis, DNA bands can be visualized by staining with ethidium bromide and exposing to UV light. Agarose gel electrophoresis is used for applications like analyzing restriction enzyme digestion products and determining DNA sizes.






![Buffer
During electrophoresis water undergoes hydrolysis :
H2O H+ + OH-
Buffers prevent the pH from changing by reacting with the H+ or OH- products
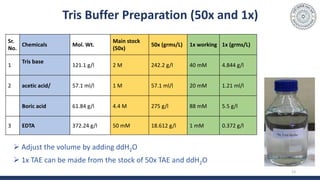
Components of buffer (TBE or TAE) used :
- TRIS [tris(hydroxymethyl)aminomethane]
- Boric Acid or acetic acid
- EDTA (Ethylenediamine tetra-acetic acid)
-for chelating the Mg2+ ions which are cofactors for DNA nucleases
12
Tris is a strong base and borate/AA is an
acid, combination of both maintains the
pH nearly 8 to 8.5](https://image.slidesharecdn.com/gelelectrophoresispractical-210515114604/85/Gel-electrophoresis-practical-12-320.jpg)