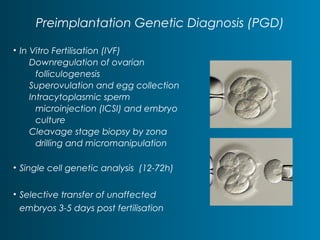
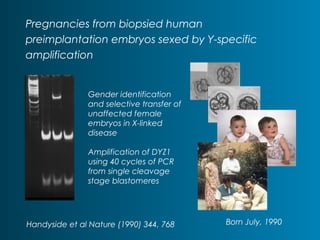
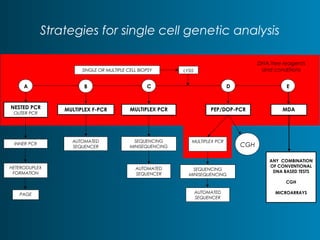
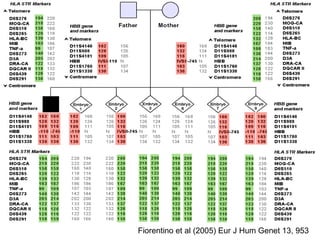
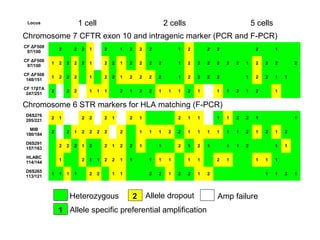
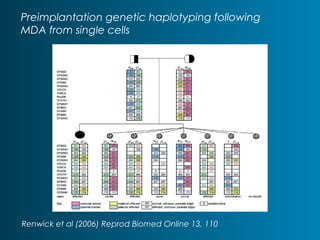
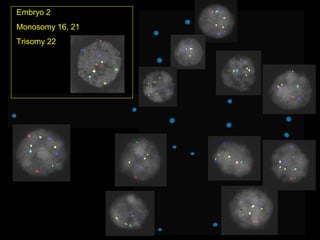
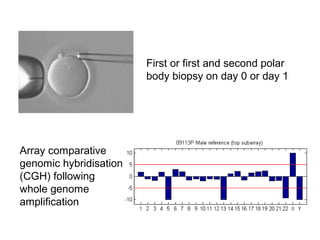
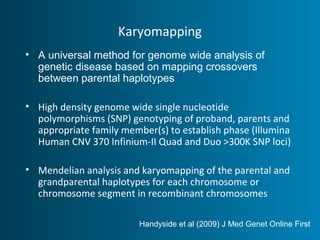
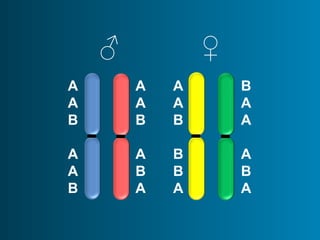
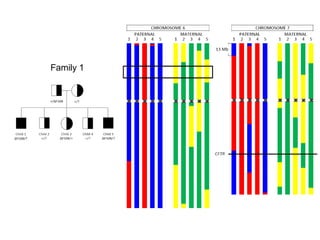
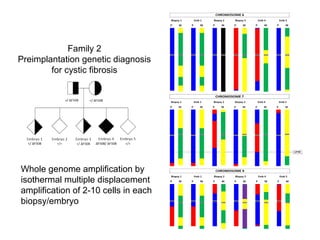
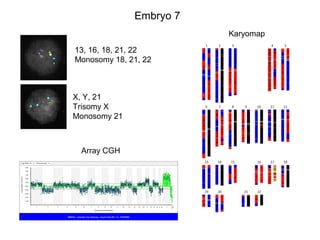
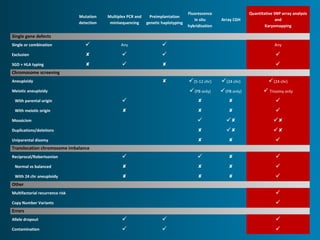
This document summarizes the history and current state of preimplantation genetic diagnosis (PGD). PGD involves in vitro fertilization, embryo biopsy, and genetic testing to select embryos without inherited diseases or chromosomal abnormalities for transfer. Over 20 years, PGD has expanded from sex selection to screening for over 100 single gene defects, chromosomal abnormalities, HLA matching, and assessing risk for multifactorial diseases. New techniques like array comparative genomic hybridization and single nucleotide polymorphism analysis with karyomapping now allow comprehensive chromosome and single gene defect screening, identification of aneuploidies, duplications/deletions, and determination of parental origins.