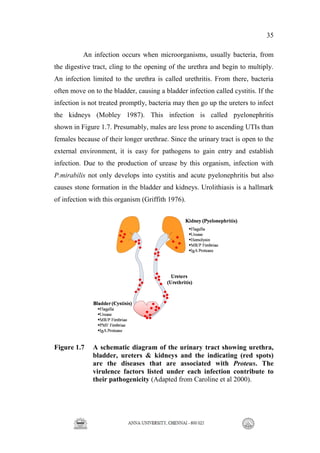
This document describes the development of a new diagnostic method called ProteAl for the rapid detection of Proteus bacteria. 2-methylbutanal was identified as a volatile organic compound biomarker specifically produced by Proteus. A fluorescent assay was developed using the reagent 5-dimethylaminonaphthalene-1-sulfonylhydrazine to detect 2-methylbutanal. This ProteAl assay could identify Proteus within 7 hours of growth and differentiated it from other common uropathogens. The production of 2-methylbutanal by Proteus was found to be regulated by the isoleucine metabolic pathway. Rational design of growth medium with increased isoleucine enhanced the yield of





























![xxx
DPO - Polymer-Deposited Optical sensors
PTR-MS - Proton-transfer-reaction mass spectrometry
qPCR - Quantitative PCR
QCM - Quartz crystal microbalance
RFU - Relative Fluorescent Unit
Rt - Retention time
RT-PCR - Reverse Transcriptase Polymerase Chain
Reaction
RNaseA - RibonucleaseA
RNA - Ribonucleic acid
rpm - Rotations per minute
SS agar - Salmonella-Shigella agar
SEB - Self-encoded bead
SDS - Sodium dodecyl sulfate
SHE - Static Headspace Extraction
SAW - Surface Acoustic Wave
TPP - Thiamine pyrophosphate
TSM - Thickness-shear mode
TSI - Triple sugar iron test
TBE - Tris Borate EDTA
Tris - Tris-[Tris-(hydroxy methyl) amino methane]
TSB - Tryptone Soya broth
UTI - Urinary Tract Infections
UPEC - Uropathogenic Escherichia coli
Val - Valine
VNC - Viable-but-nonculturable
VOCs - Volatile Organic Compounds
WBCs - White blood cells
WHO - World Health Organization](https://image.slidesharecdn.com/fec77b75-c103-44ff-be5b-f15dd974cc93-160613124202/85/Full-Thesis-30-320.jpg)





























































![63
Table 2.5 List of environmental sample collection locations
Location Type of waste dumped
Madipakkam, Chennai Hospital waste
Pallikaranai, Chennai Domestic waste
Taramani, Chennai Laboratory waste
Around 10 g of soil samples were collected from each location by
digging the ground approximately 6 inches below the surface. From this 1 g
of the soil was dissolved in 10 ml of sterile distilled water and was serially
diluted to ~105
cells and plated onto LB agar plates. Morphologically different
colonies were isolated and common microbiological and biochemical tests
were performed followed by screening with ProteAl assay.
2.11 SENSITIVITY AND SPECIFICITY CALCULATION
Sensitivity and specificity of the assay was calculated using the
formula, Sensitivity = [a/ (a+c)] ×100 and Specificity = [d/ (b+d)] ×100
Table 2.6 Table for sensitivity and specificity calculation
True positive (a) False positive (b)
False negative (c) True negative (d)
When the growth (OD) of the strains where similar, the 99 %
confidence for the positive (Proteus) and negatives were calculated using the
formula.](https://image.slidesharecdn.com/fec77b75-c103-44ff-be5b-f15dd974cc93-160613124202/85/Full-Thesis-93-320.jpg)
















































































![146
LIST OF PUBLICATIONS
1. Raju Aarthi, Raju Saranya & Krishnan Sankaran 2014,
‘2-methylbutanal, a volatile biomarker, for non-invasive surveillance
of Proteus’, Appl Microbiol. Biotechnology, vol. 98, no.1, pp.445-454.
2. Raju Saranya, Raju Aarthi & Krishnan Sankaran 2015, ‘Simple and
specific colorimetric detection of Staphylococcus using its volatile
2-[3-acetoxy-4,4,14-trimethylandrost-8-en-17-yl] propanoic acid in the
liquid phase and head space of cultures’, Appl Microbiol
Biotechnology, vol. 99, no. 10, pp. 4423-33.](https://image.slidesharecdn.com/fec77b75-c103-44ff-be5b-f15dd974cc93-160613124202/85/Full-Thesis-176-320.jpg)