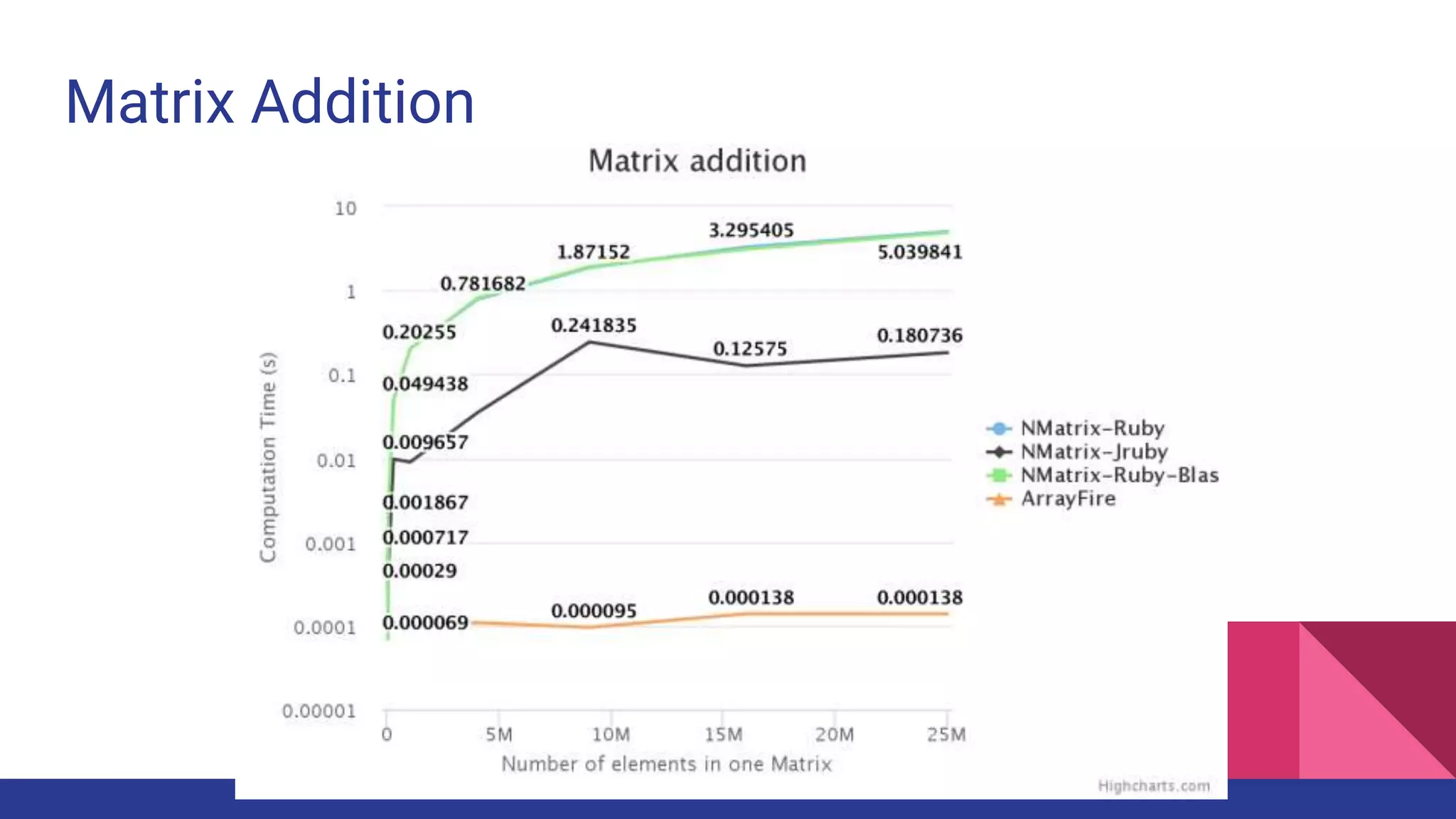
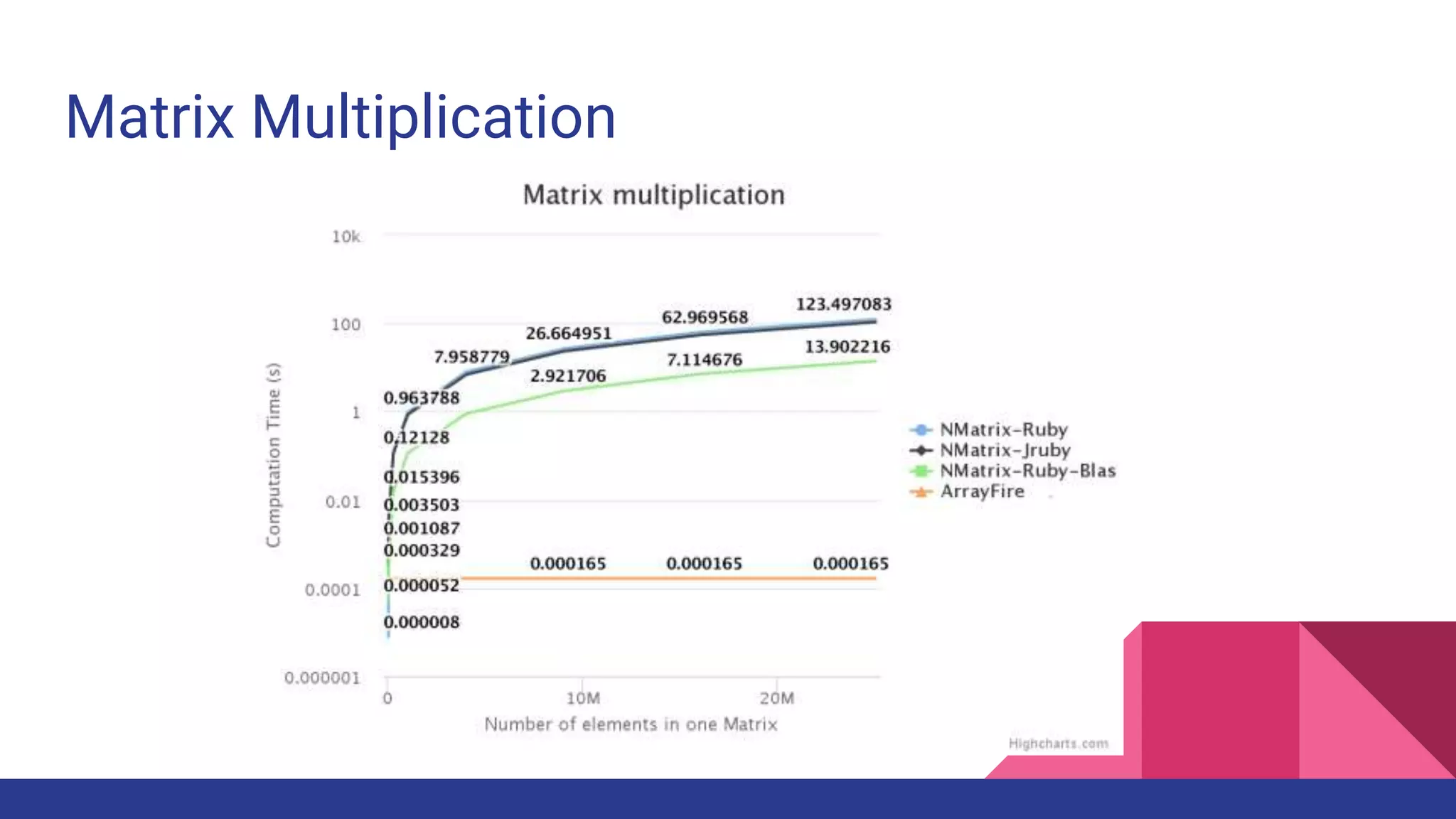
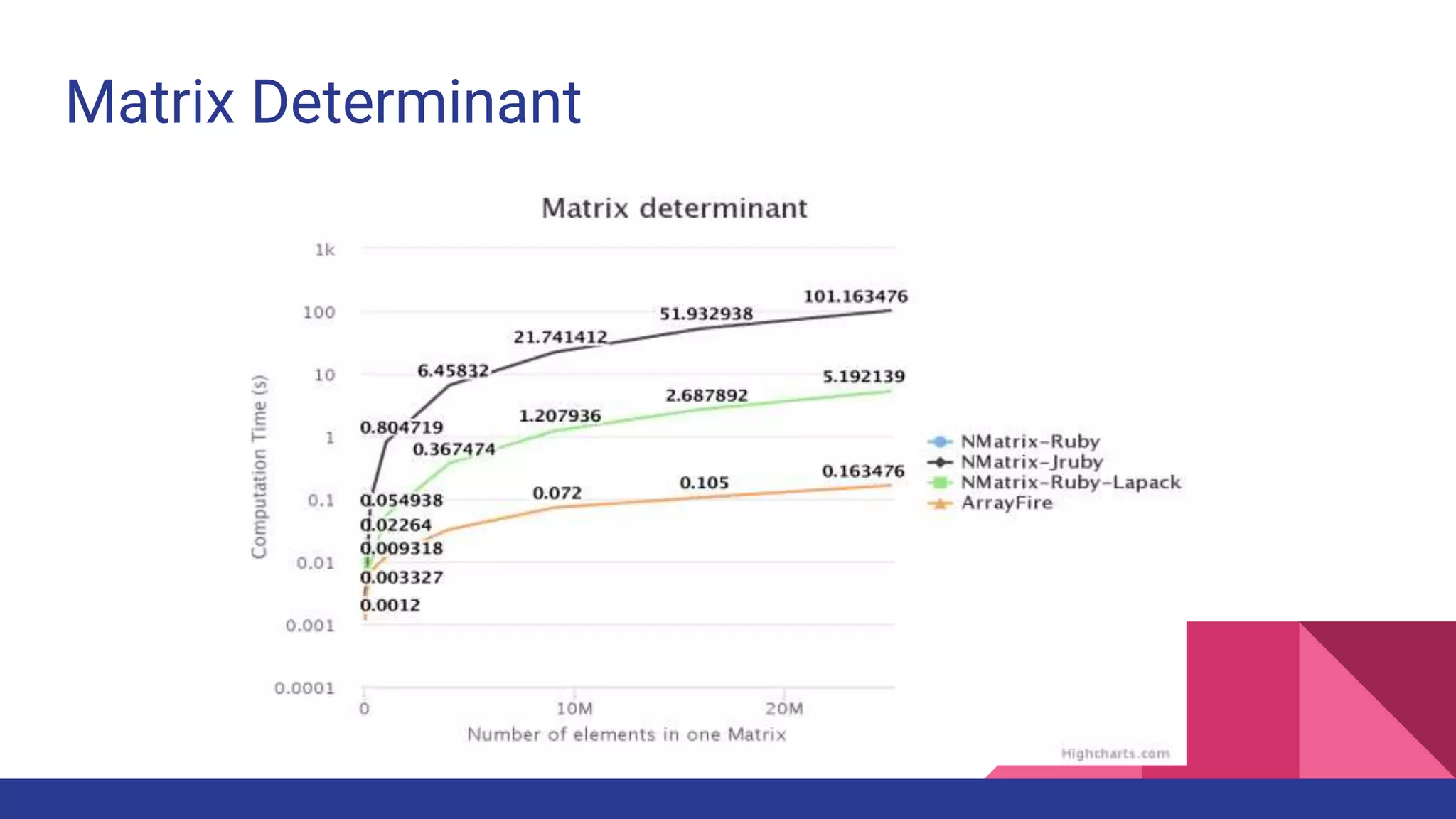
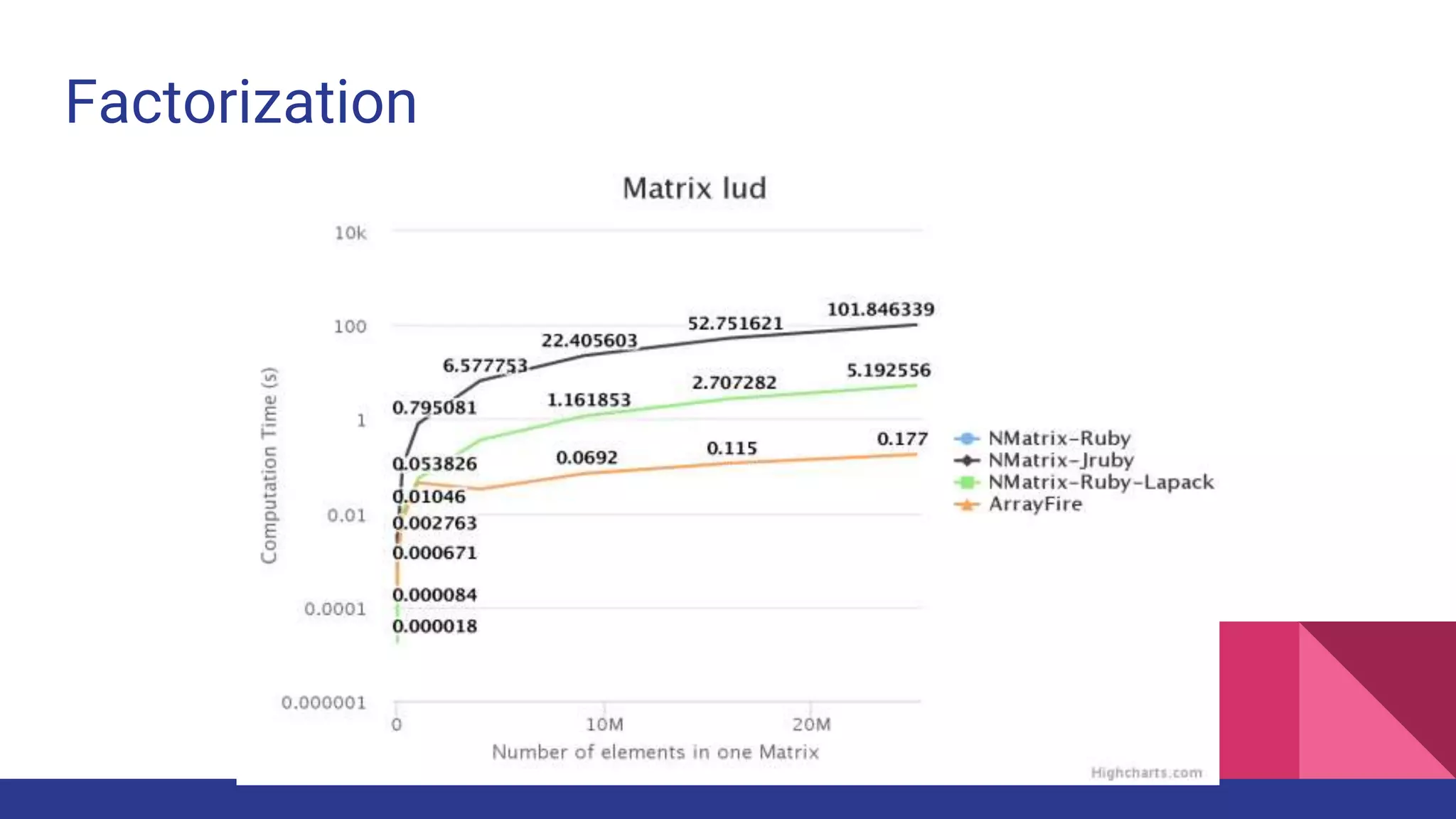
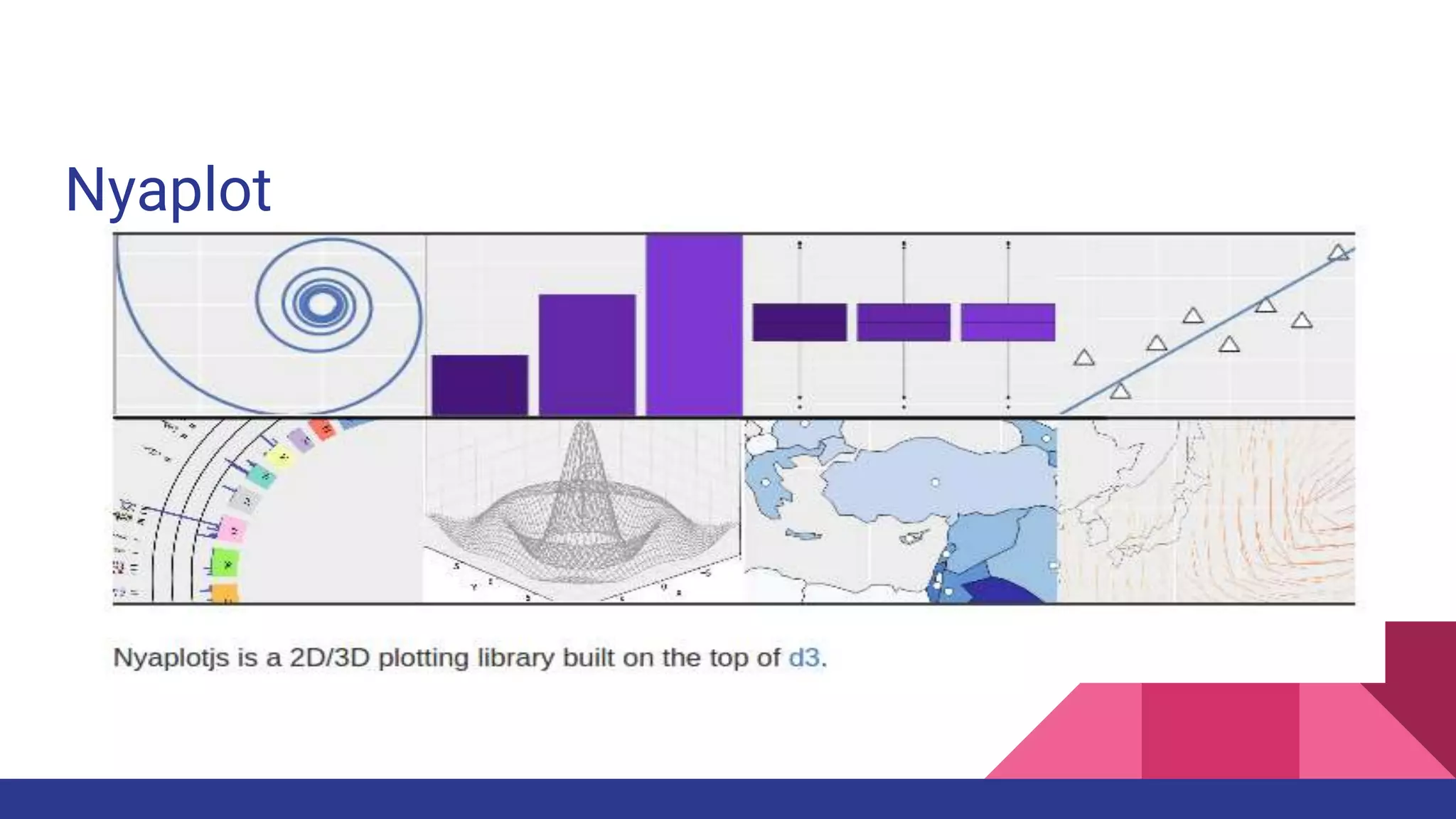
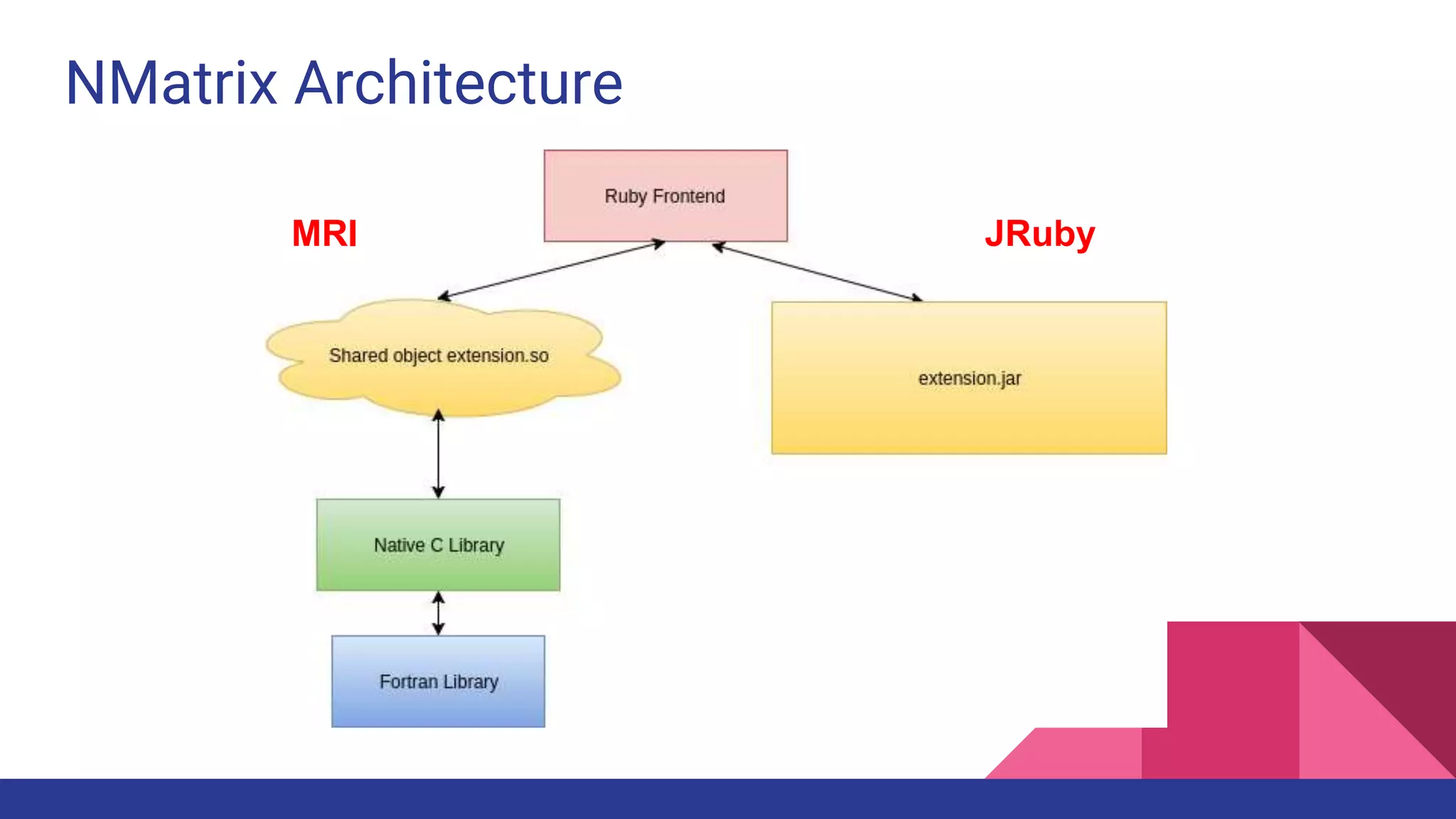
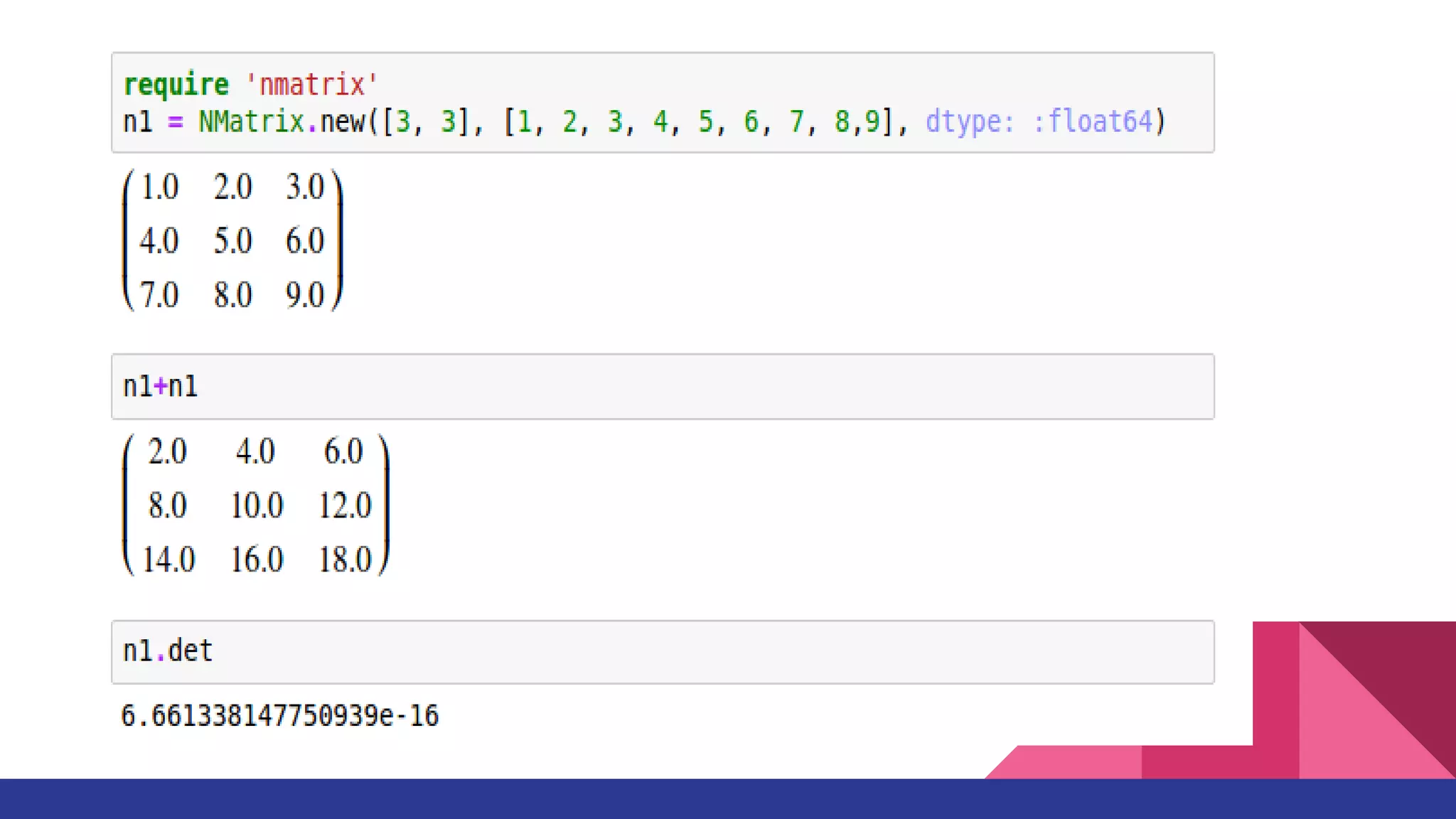
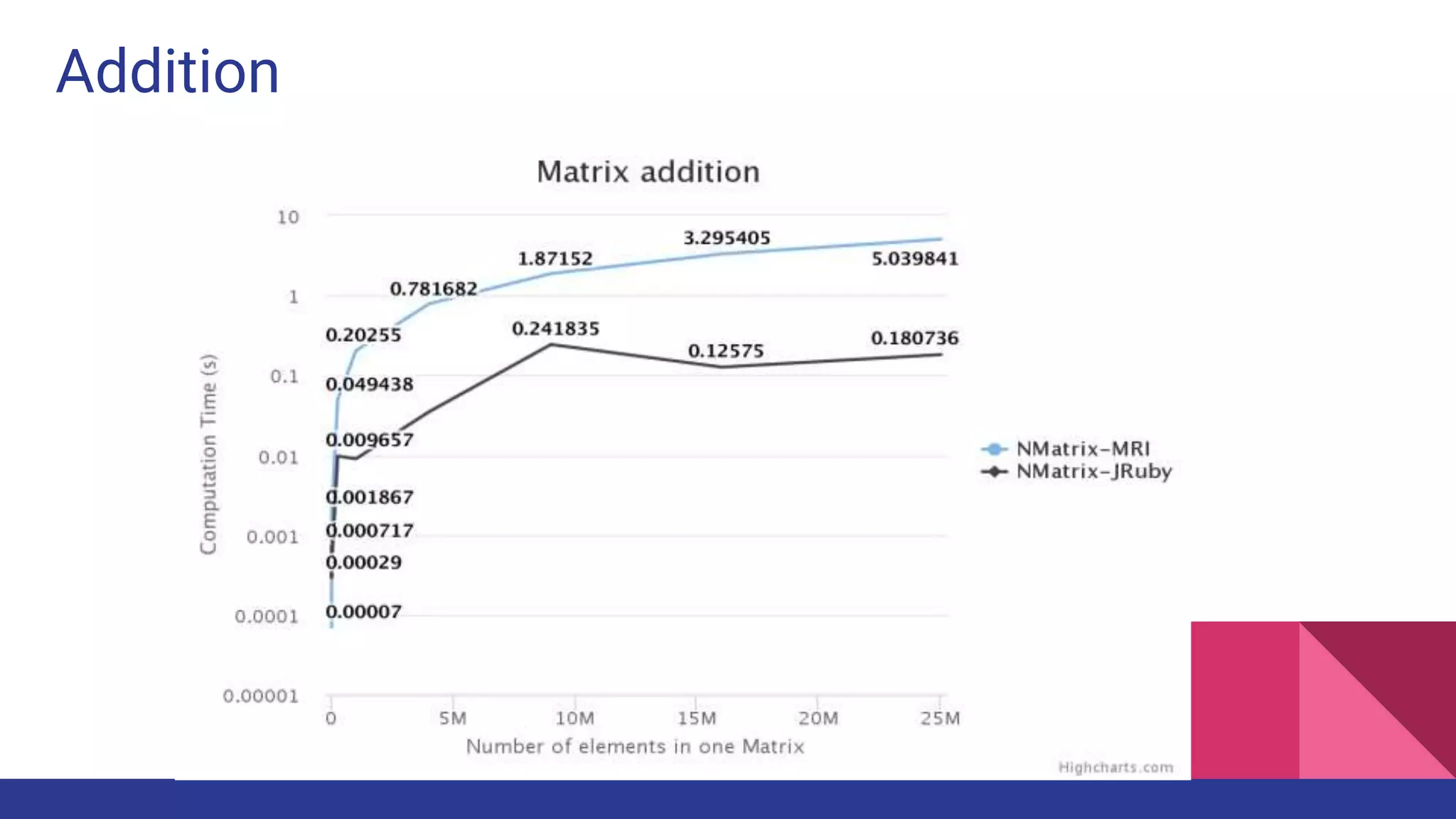
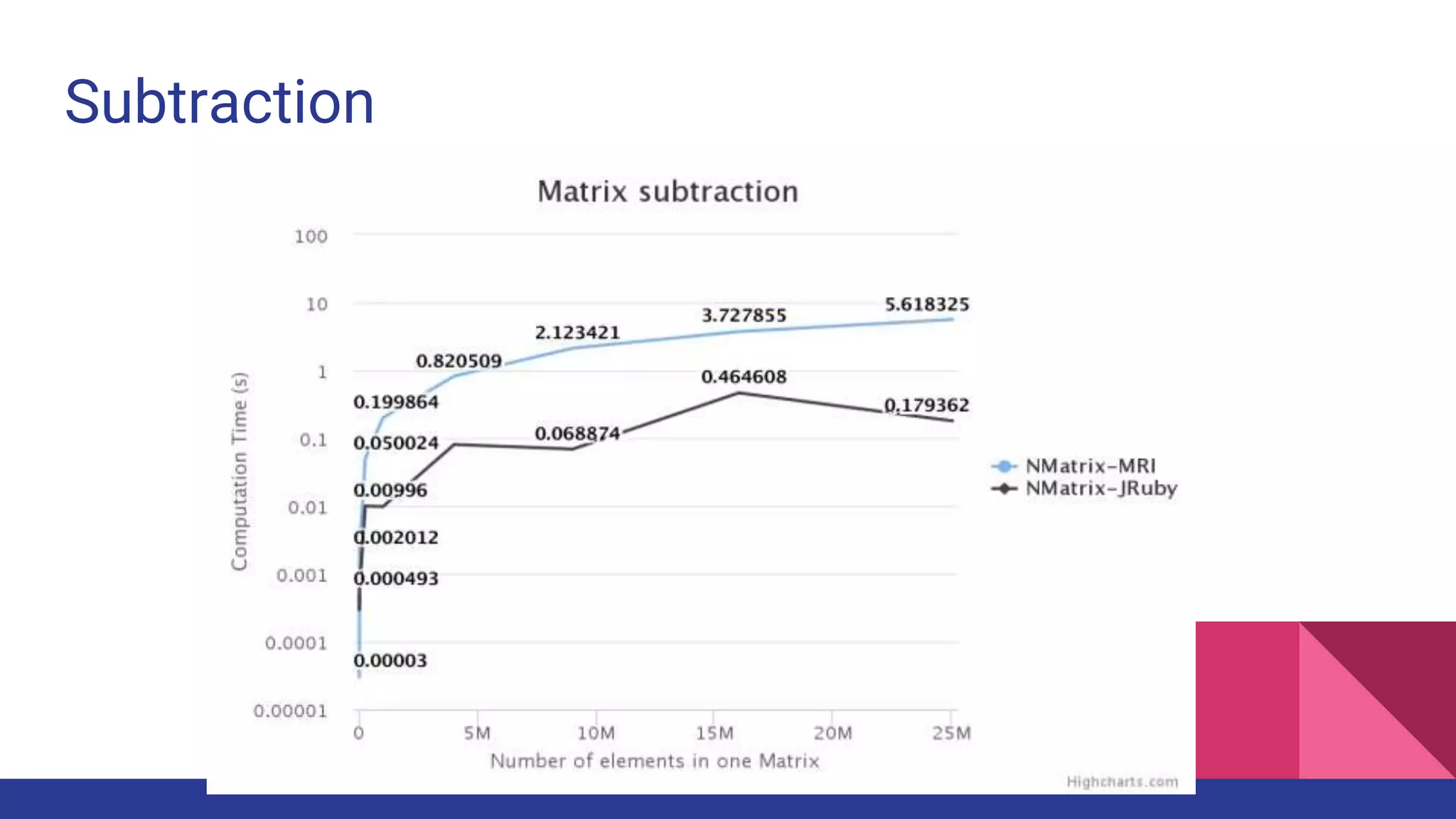
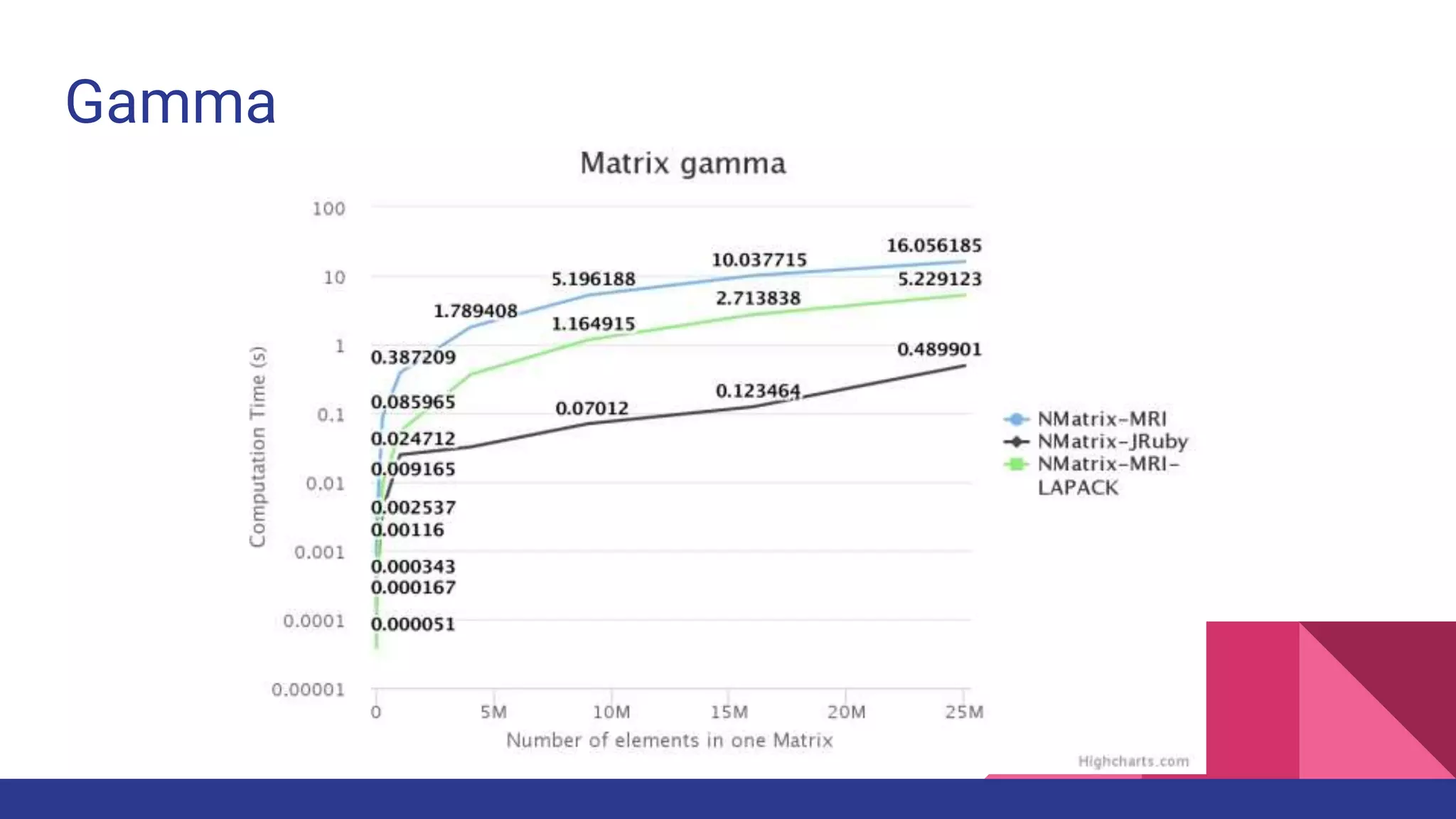
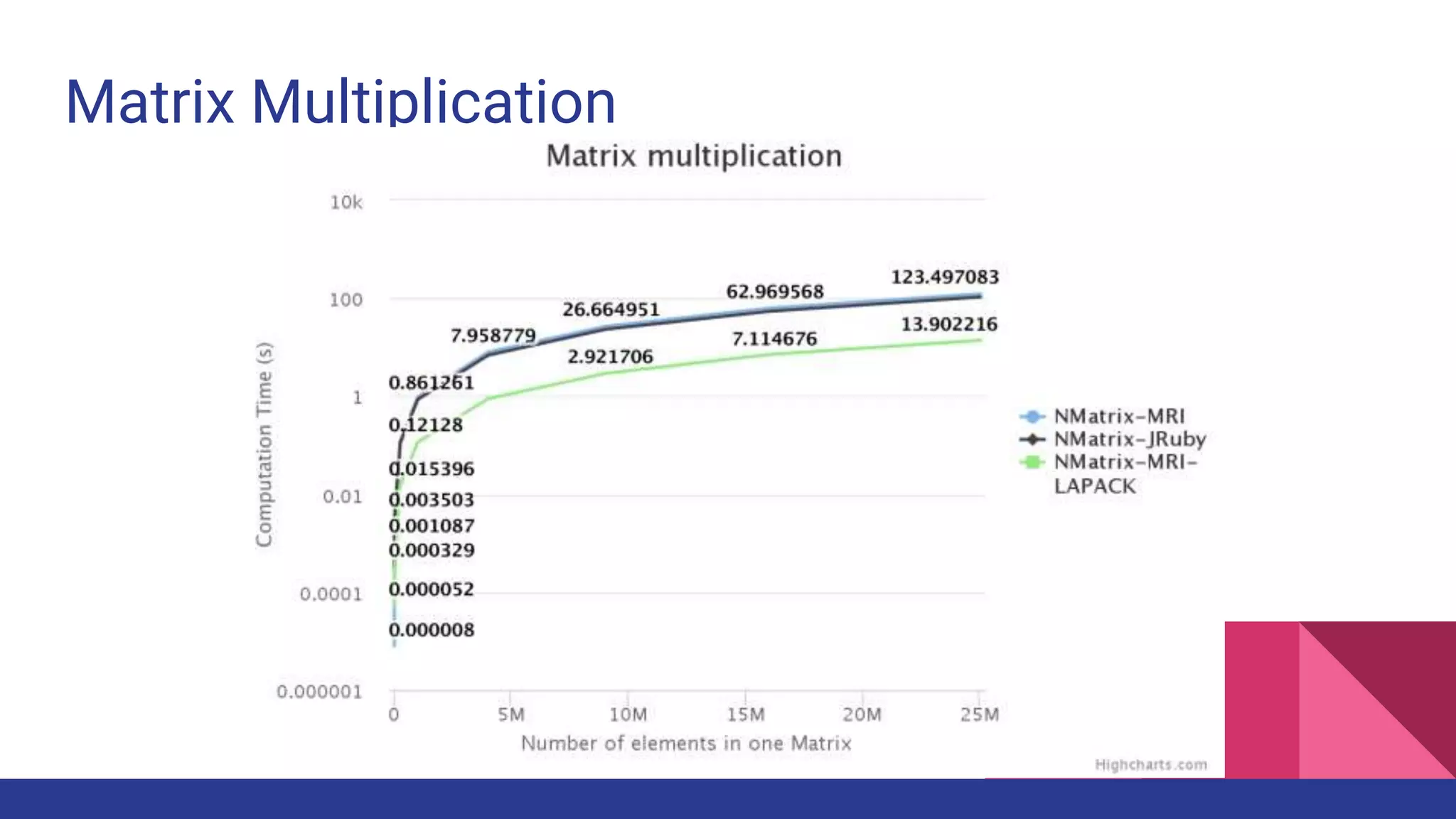
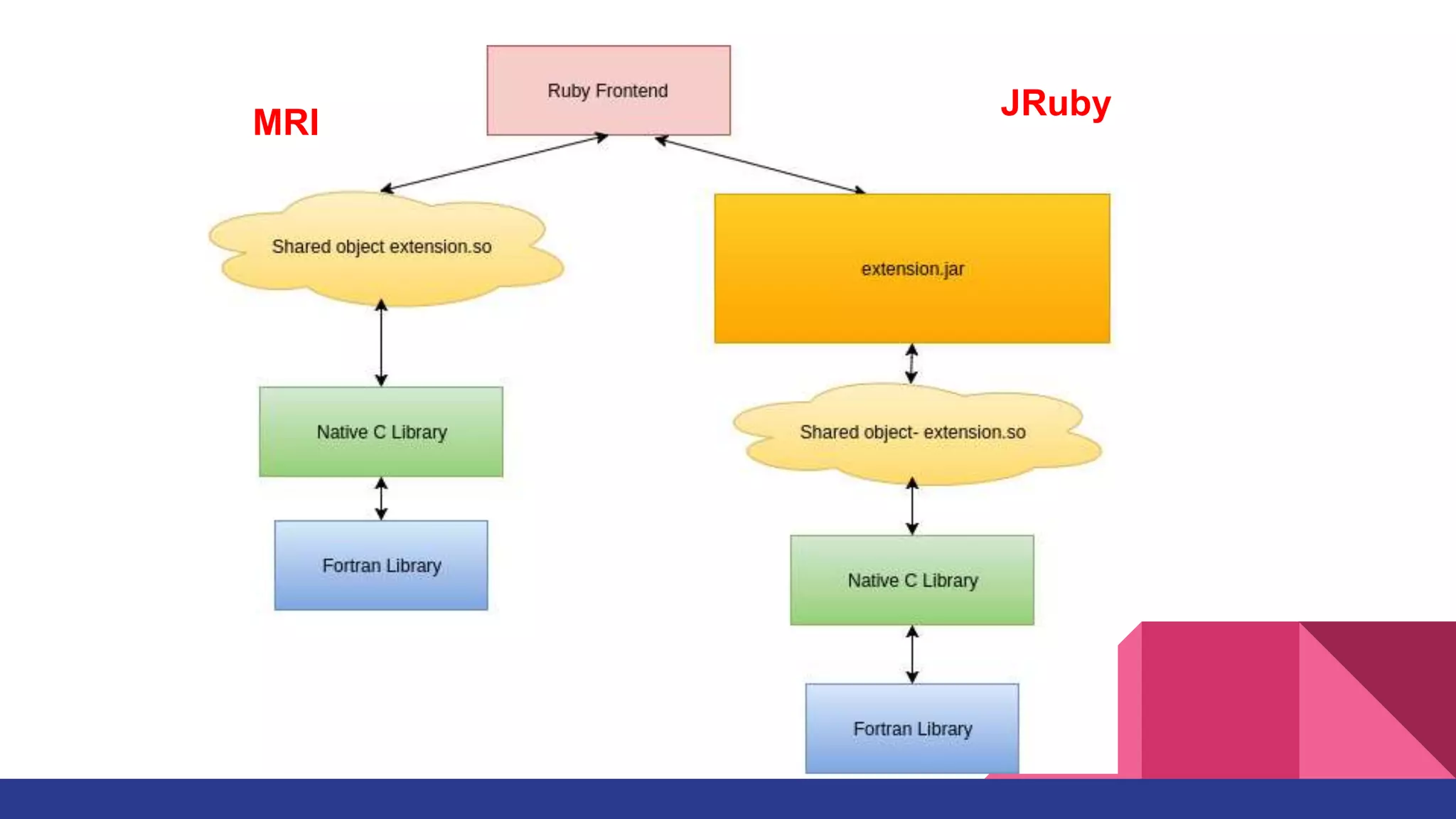
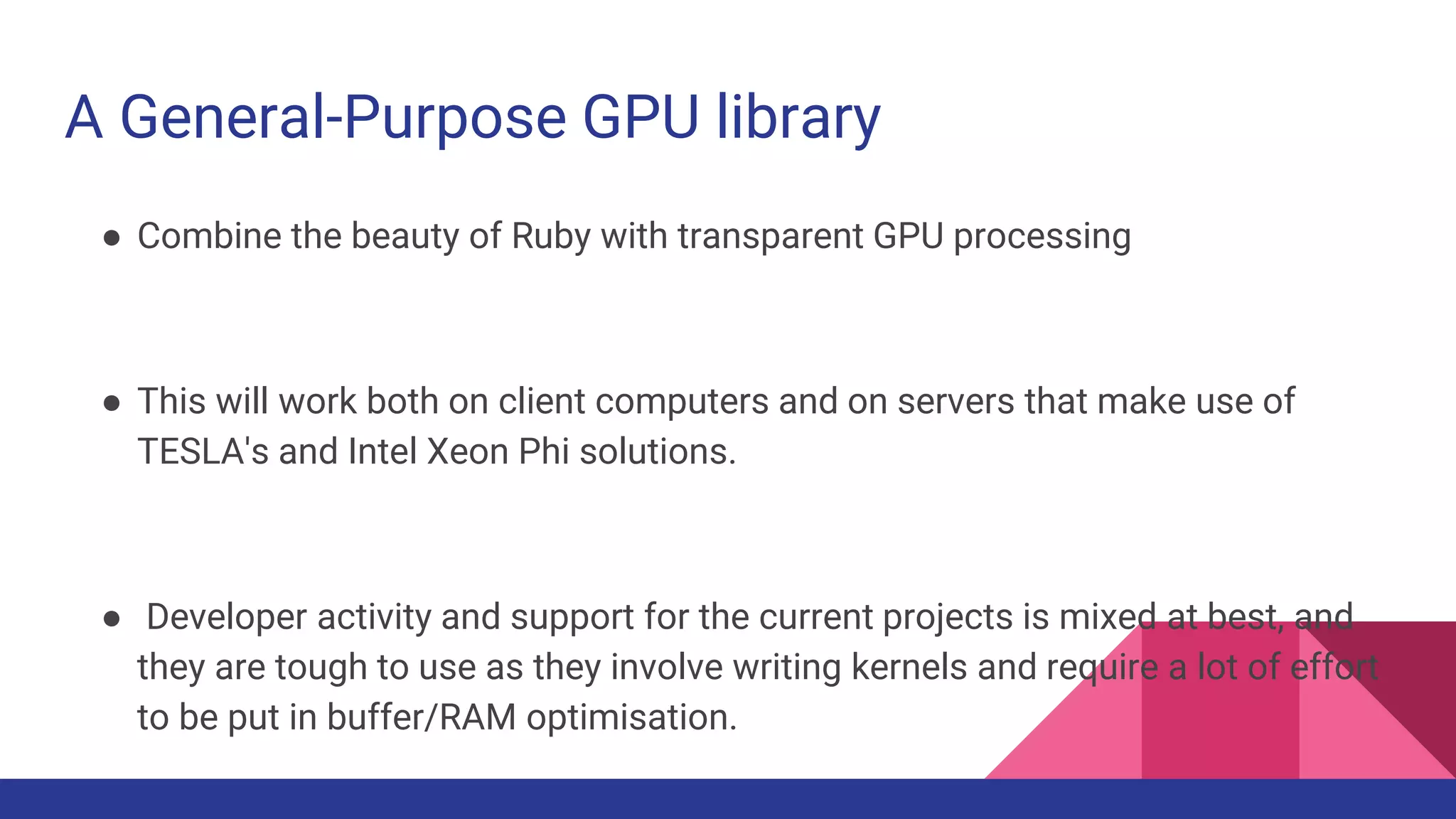
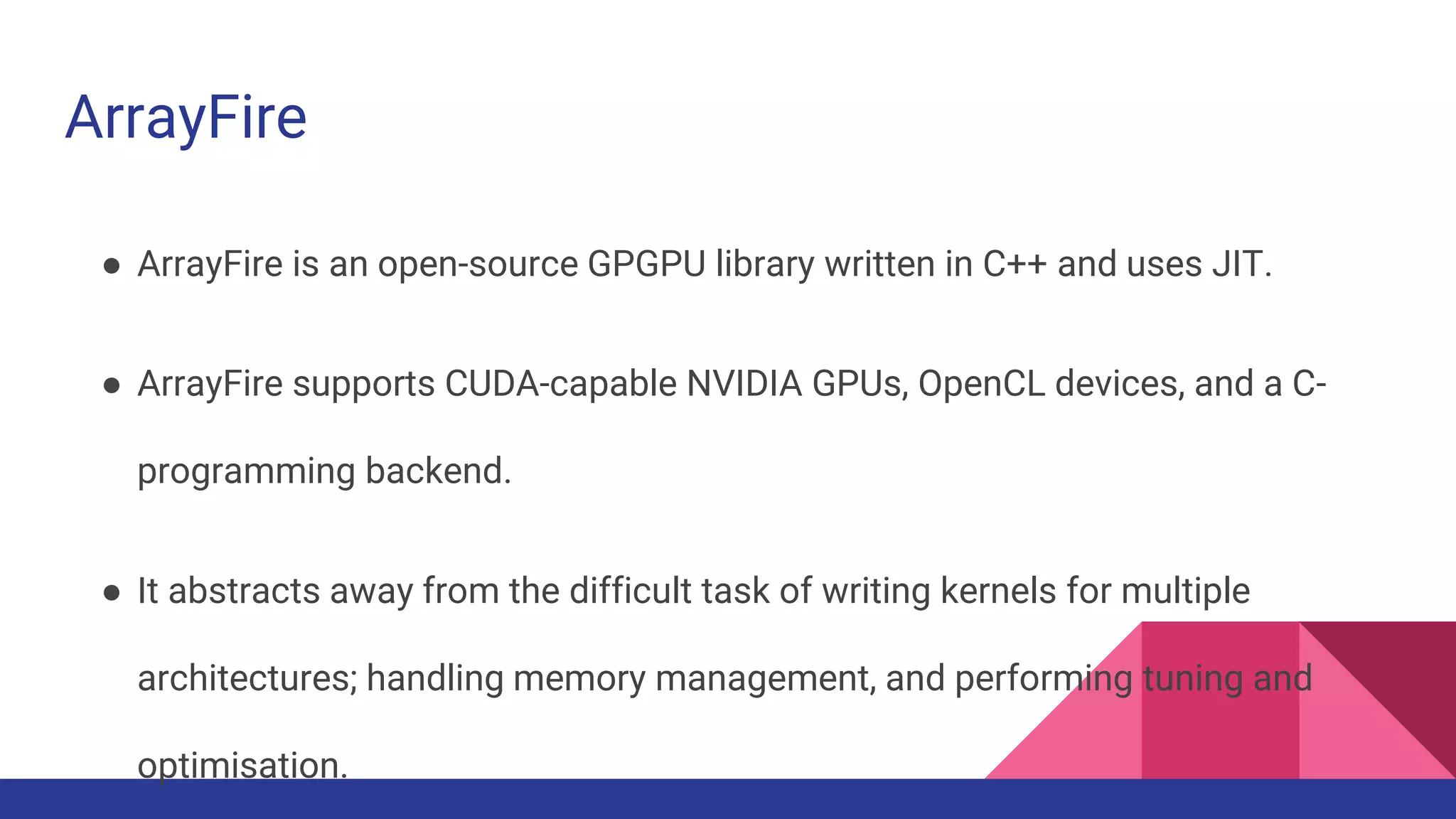
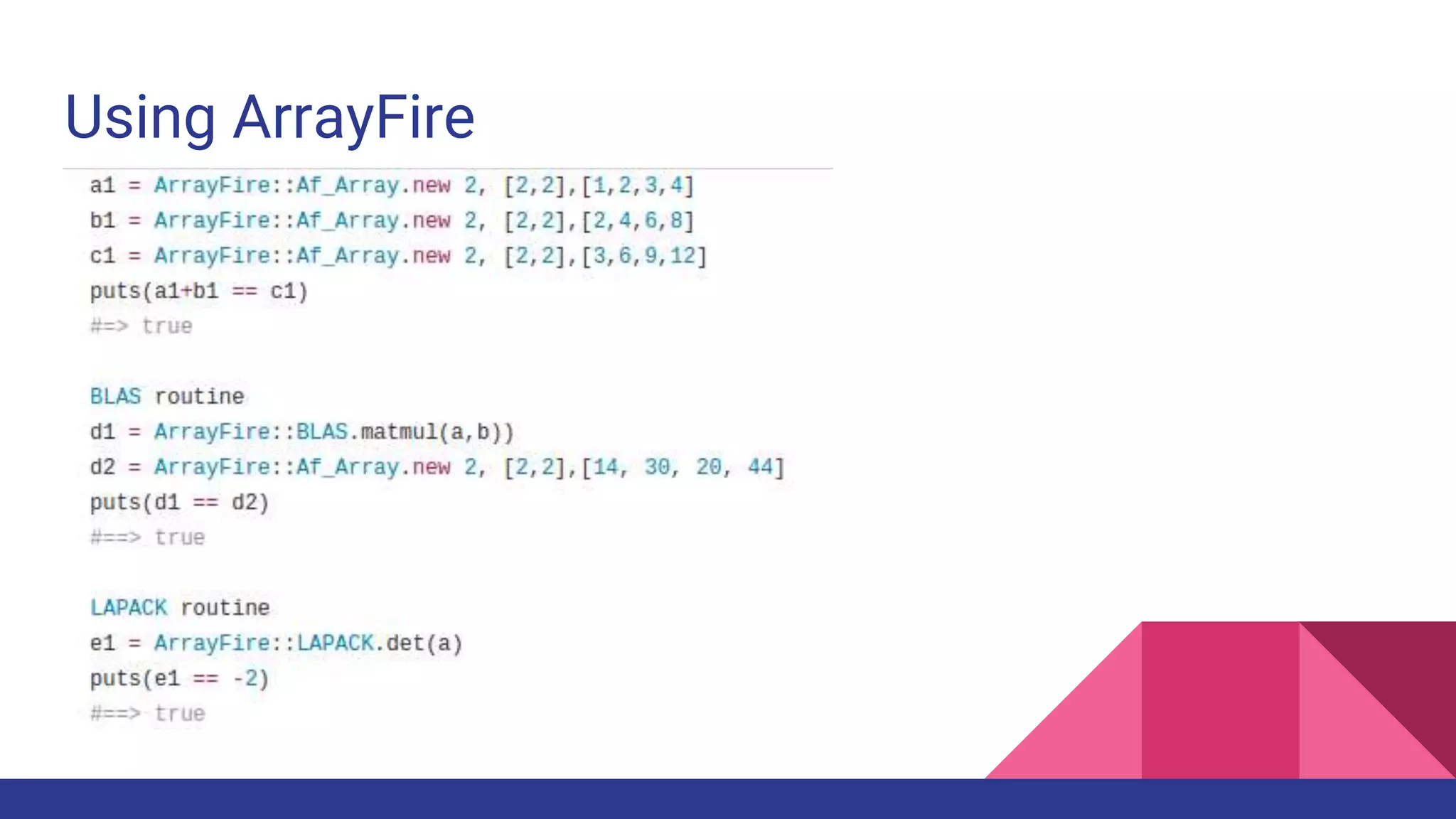
Scientific computing in Ruby is advancing with libraries like NMatrix, Daru, and Mixed_models. NMatrix provides numerical matrix functionality and initially relied on ATLAS/CBLAS for operations, but a JRuby version using Apache Commons Math is much faster. A proposed general purpose GPU library called ArrayFire-rb would combine Ruby with transparent GPU processing using the ArrayFire C++ library. It could be implemented for MRI using C extensions inspired by NMatrix and for JRuby using Java Native Interface. Benchmarking shows the JRuby version of NMatrix is vastly faster for elementwise operations on n-dimensional matrices compared to MRI. ArrayFire integration could further accelerate applications in fields like bioinformatics, image processing, and computational fluid dynamics






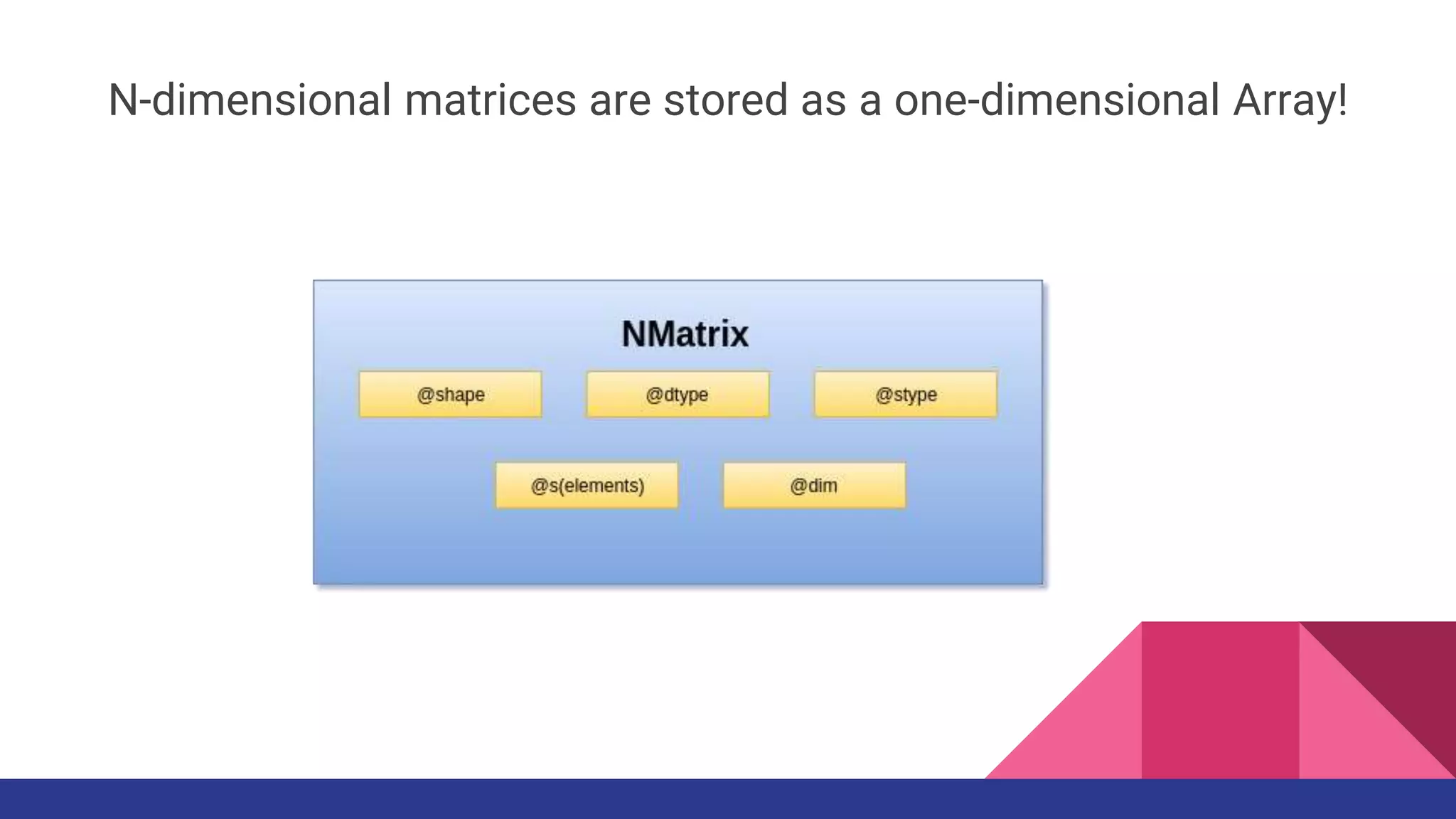

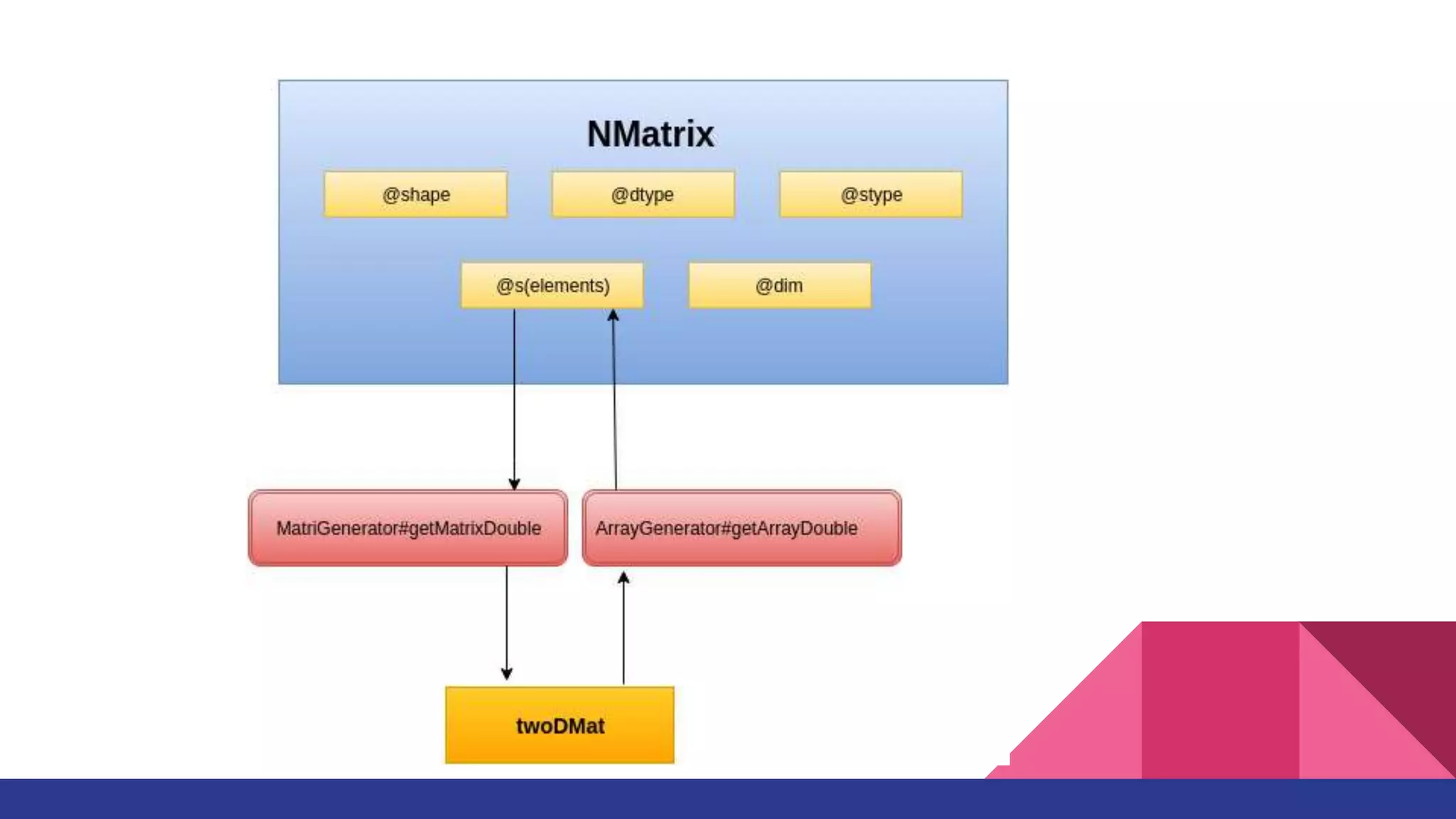
![Elementwise Operation
● [:add, :subtract, :sin, :gamma]
● Iterate through the elements.
● Access the element; do the operation, return it](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-18-2048.jpg)

![Errors that can’t be reproduced :p
[ 0.11, 0.05, 0.34, 0.14 ]
+ [ 0. 21, 0.05, 0.14, 0.14 ]
= [ 0, 0, 0, 0]
([ 0. 11, 0.05, 0.34, 0.14 ] + 5)
+ ([ 0. 21, 0.05, 0.14, 0.14 ] + 5)
- 10
= [ 0.32, 0.1, 0.48, 0.28]](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-21-2048.jpg)
![Autoboxing
● :float64 => double only
● Strict dtypes => creating data type in Java. Can’t Rely on Reflection
● @s = Array.new()
● @s = Java::double[rows*cols].new()](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-22-2048.jpg)
![Autoboxing and Enumerators
def each_with_indices
nmatrix = create_dummy_nmatrix
stride = get_stride(self)
offset = 0
coords = Array.new(dim){ 0 }
shape_copy = Array.new(dim)
(0...size).each do |k|
dense_storage_coords(nmatrix, k, coords,
stride, offset)
slice_index =
dense_storage_pos(coords,stride)
ary = Array.new
if (@dtype == :object)
ary << self.s[slice_index]
else
ary << self.s.toArray.to_a[slice_index]
end
(0...dim).each do |p|
ary << coords[p]
end
yield(ary)
end if block_given?
return nmatrix
end](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-23-2048.jpg)


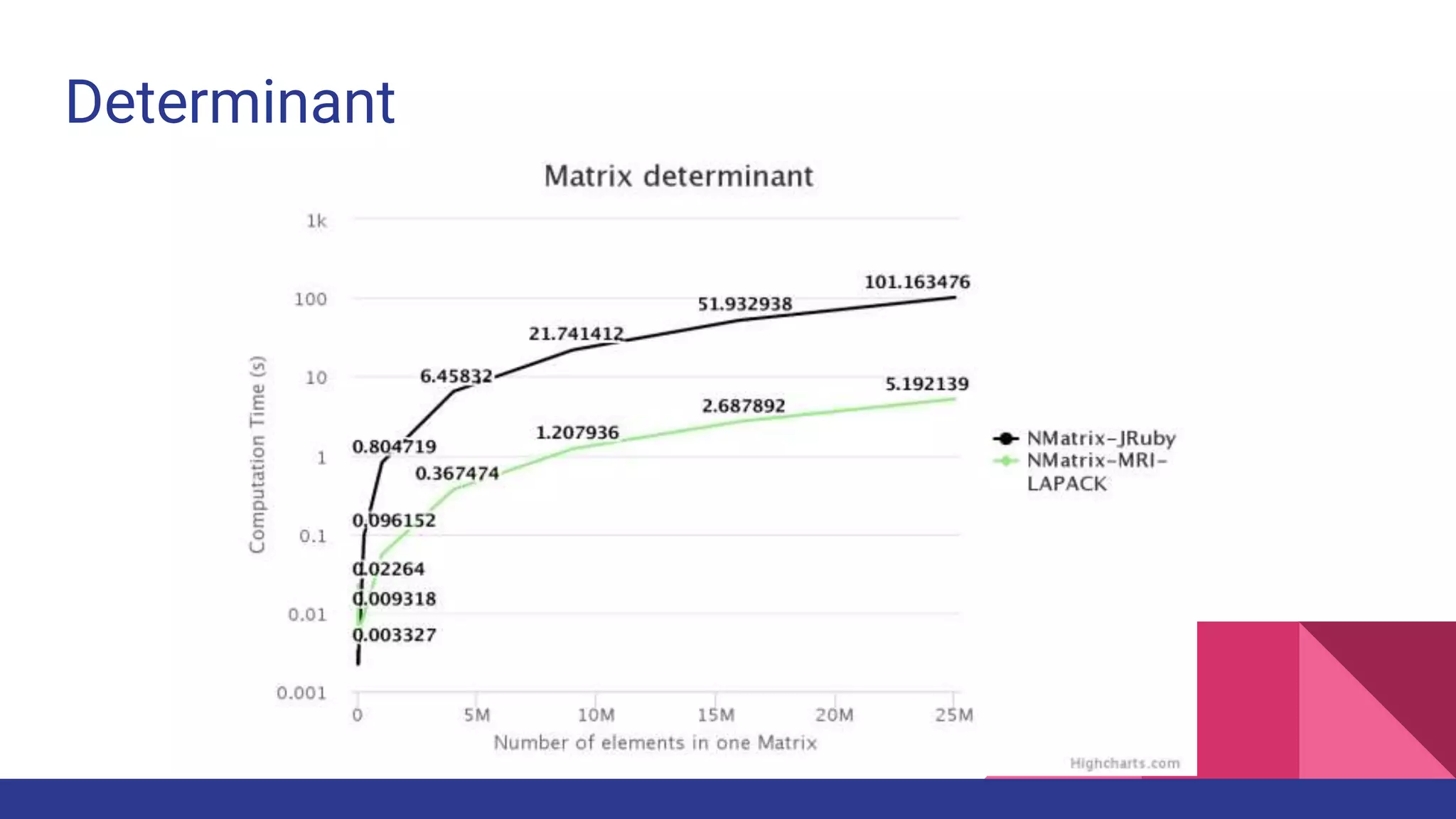
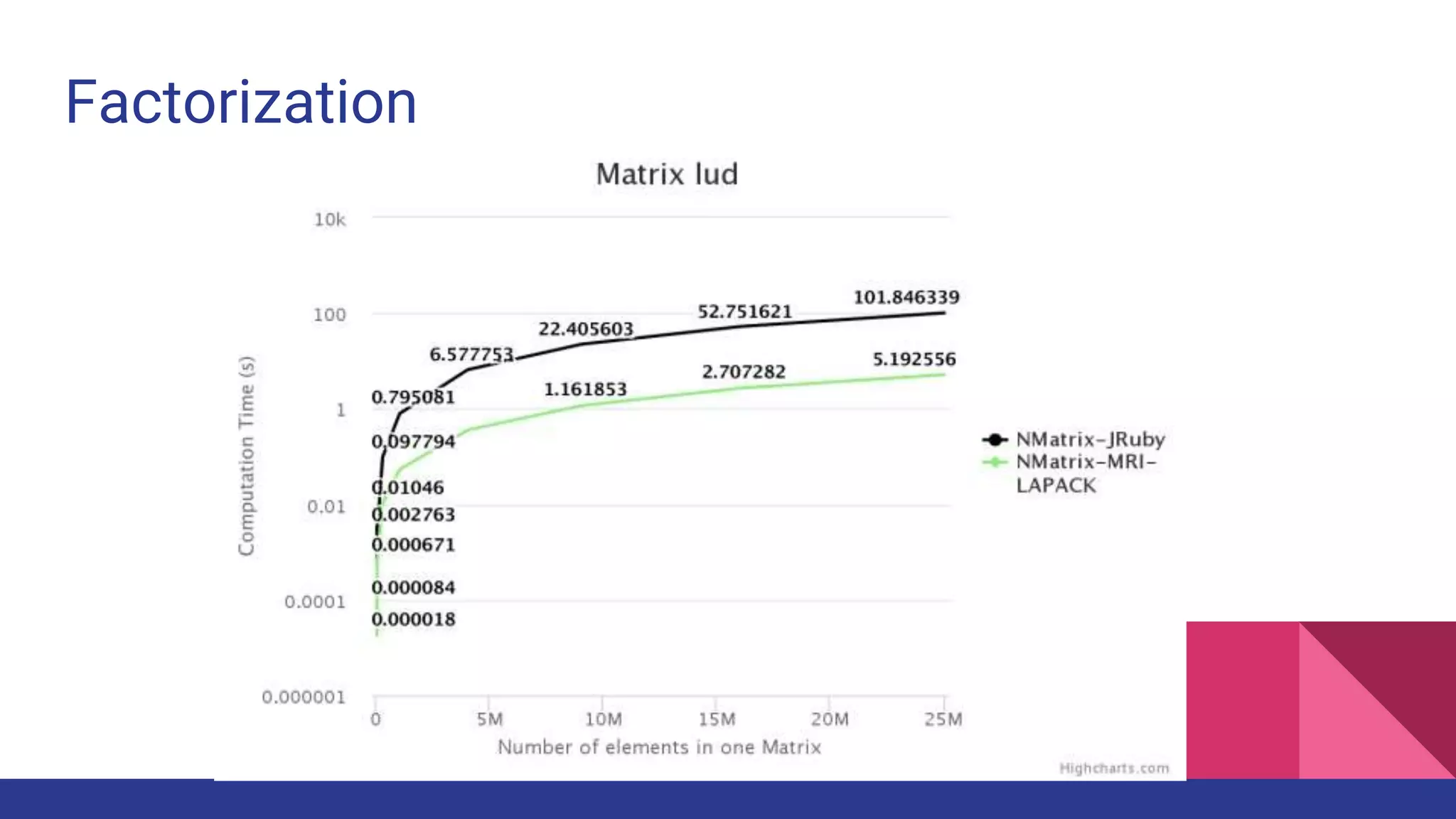
![2 - dimensional Matrix Operations
● [:dot, :det, :factorize_lu]
● In NMatrix-MRI, BLAS-III and LAPACK routines are implemented using their
respective libraries.
● NMatrix-JRuby depends on Java functions.](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-26-2048.jpg)

![Ruby Code
index =0
puts Benchmark.measure{
(0...15000).each do |i|
(0...15000).each do |j|
c[i][j] = b[i][j]
index+=1
end
end
}
#67.790000 0.070000 67.860000 ( 65.126546)
#RAM consumed => 5.4GB
b = Java::double[15_000,15_000].new
c = Java::double[15_000,15_000].new
index=0
puts Benchmark.measure{
(0...15000).each do |i|
(0...15000).each do |j|
b[i][j] = index
index+=1
end
end
}
#43.260000 3.250000 46.510000 ( 39.606356)](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-29-2048.jpg)

![Java Code
public class MatrixGenerator{
public static void test2(){
for (int index=0, i=0; i < row ; i++){
for (int j=0; j < col; j++){
c[i][j]= b[i][j];
index++;
}
}
}
puts Benchmark.measure{MatrixGenerator.test2}
#0.034000 0.001000 00.034000 ( 00.03300)
#RAM consumed => 300MB
public class MatrixGenerator{
public static void test1(){
double[][] b = new double[15000][15000];
double[][] c = new double[15000][15000];
for (int index=0, i=0; i < row ; i++){
for (int j=0; j < col; j++){
b[i][j]= index;
index++;
}
}
}
puts Benchmark.measure{MatrixGenerator.test1}
#0.032000 0.001000 00.032000 ( 00.03100)](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-31-2048.jpg)












![#include <ruby.h>
typedef struct AF_STRUCT
{
size_t ndims;
size_t count;
size_t* dimension;
double* array;
}afstruct;
void Init_arrayfire() {
ArrayFire = rb_define_module("ArrayFire");
Blas = rb_define_class_under(ArrayFire, "BLAS",
rb_cObject);
rb_define_singleton_method(Blas, "matmul",
(METHOD)arf_matmul, 2);
}
static VALUE arf_matmul(VALUE self, VALUE left_val, VALUE
right_val){
afstruct* left;
afstruct* right;
afstruct* result = ALLOC(afstruct);
Data_Get_Struct(left_val, afstruct, left);
Data_Get_Struct(right_val, afstruct, right);
result->ndims = left->ndims;
size_t dimension[2];
dimension[0] = left->dimension[0];
dimension[1] = right->dimension[1];
size_t count = dimension[0]*dimension[1];
result->dimension = dimension;
result->count = count;
arf::matmul(result, left, right);
return Data_Wrap_Struct(CLASS_OF(left_val), NULL,
arf_free, result);
}](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-54-2048.jpg)
![#include <arrayfire.h>
namespace arf {
using namespace af;
static void matmul(afstruct *result, afstruct *left, afstruct *right)
{
array l = array(left->dimension[0], left->dimension[1], left->array);
array r = array(right->dimension[0], right->dimension[1], right->array);
array res = matmul(l,r);
result->array = res.host<double>();
}
}
extern "C" {
#include "arrayfire.c"
}](https://image.slidesharecdn.com/fosdem2017-scientificcomputingonjruby-170204173624/75/Fosdem2017-Scientific-computing-on-Jruby-55-2048.jpg)