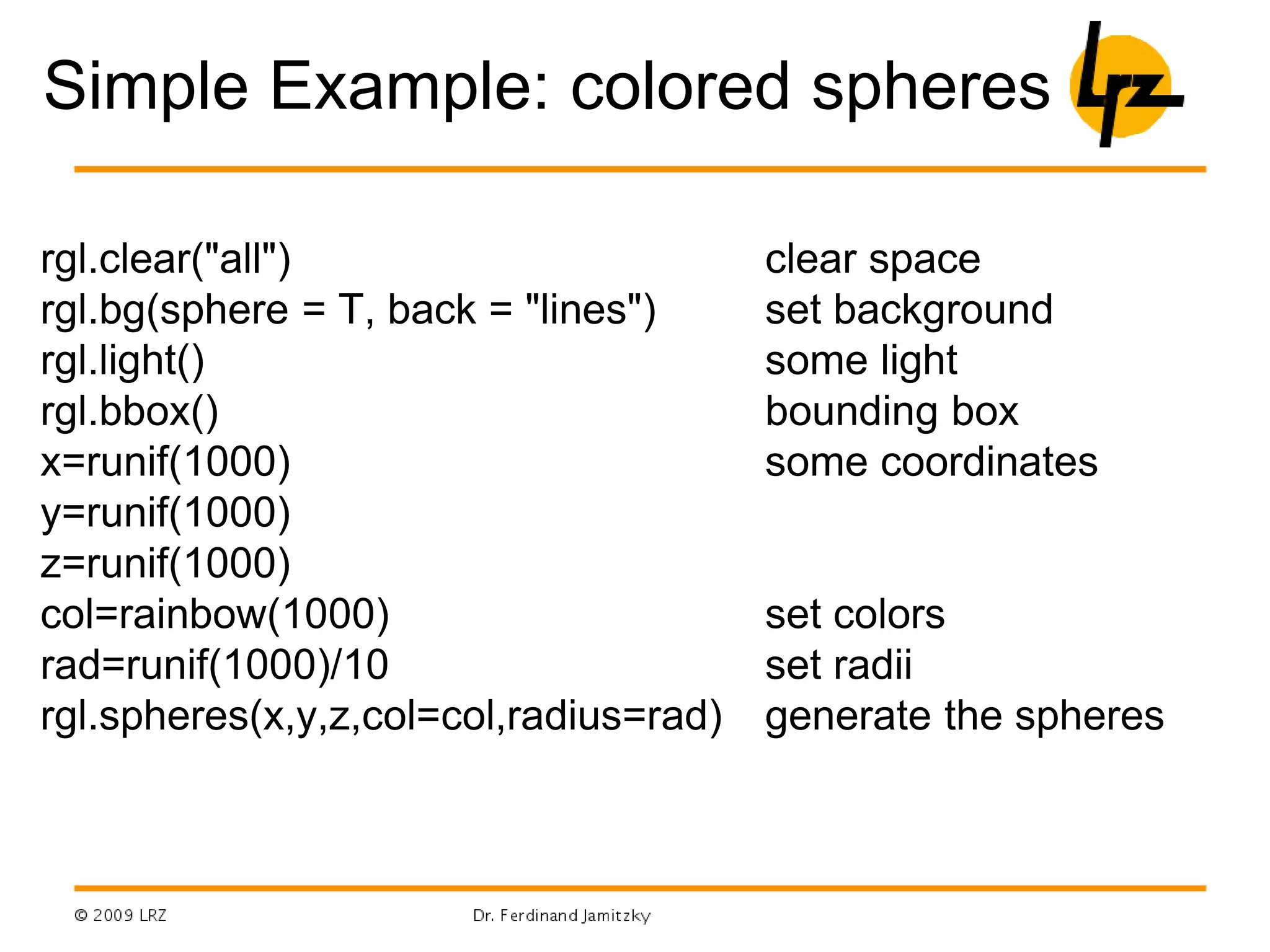
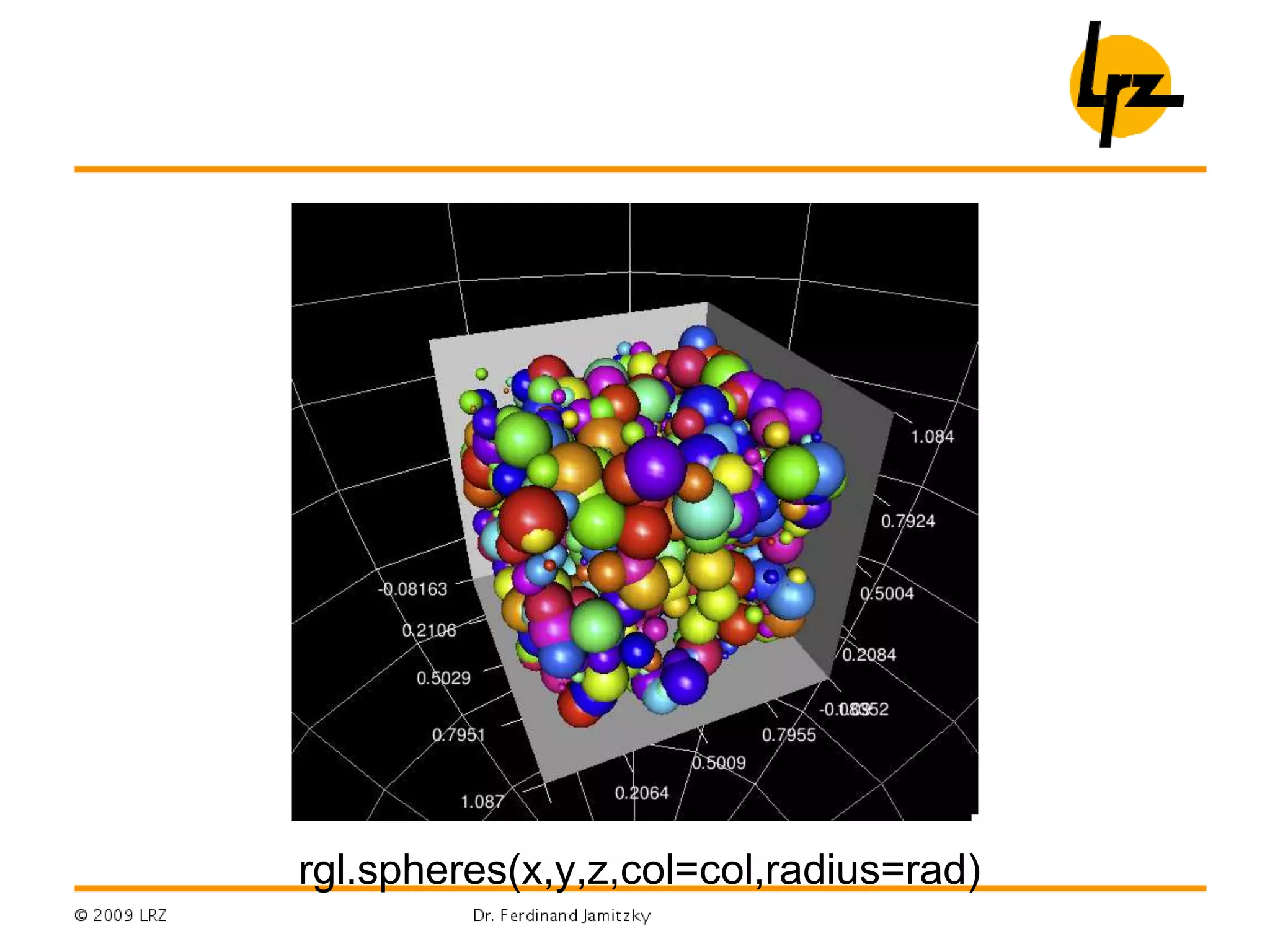
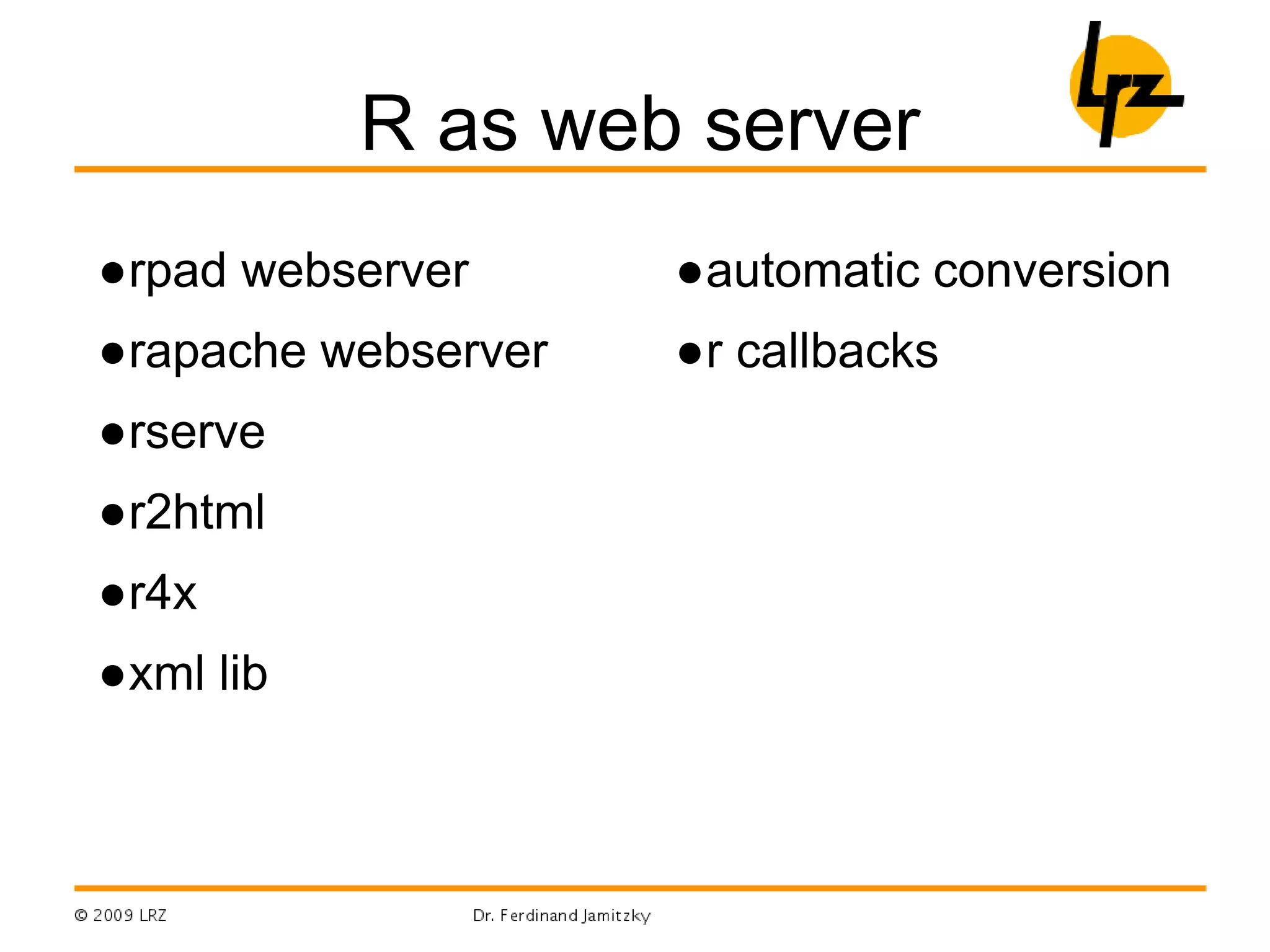
R can be used for a wide range of tasks including:
- Summarizing, R can integrate with C/C++, Fortran, Java and other languages and APIs like OpenGL, MPI, and web services.
- Binding different languages together, R objects can be passed between R and other languages like C/C++ using .C() and Fortran using .Fortran().
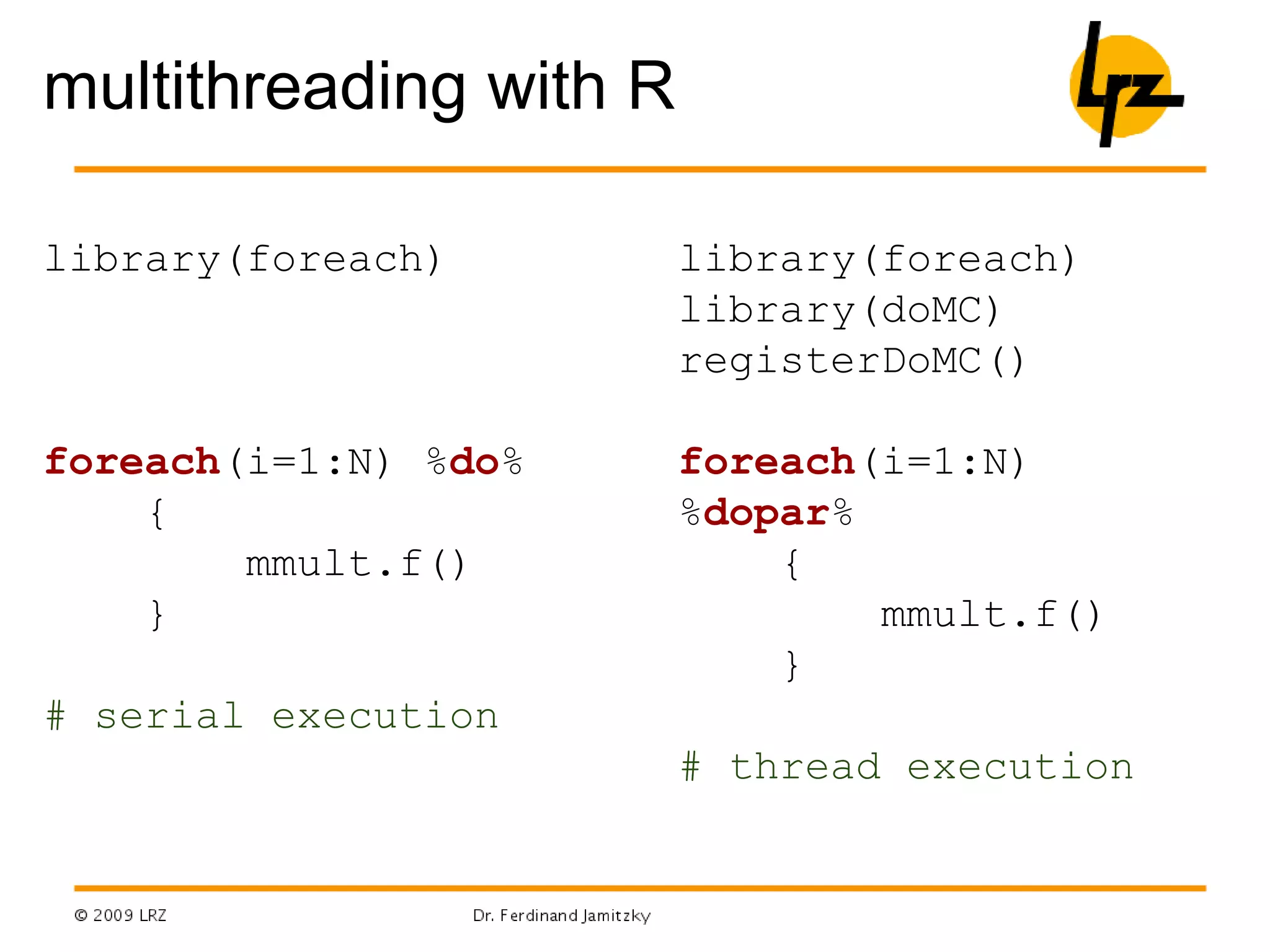
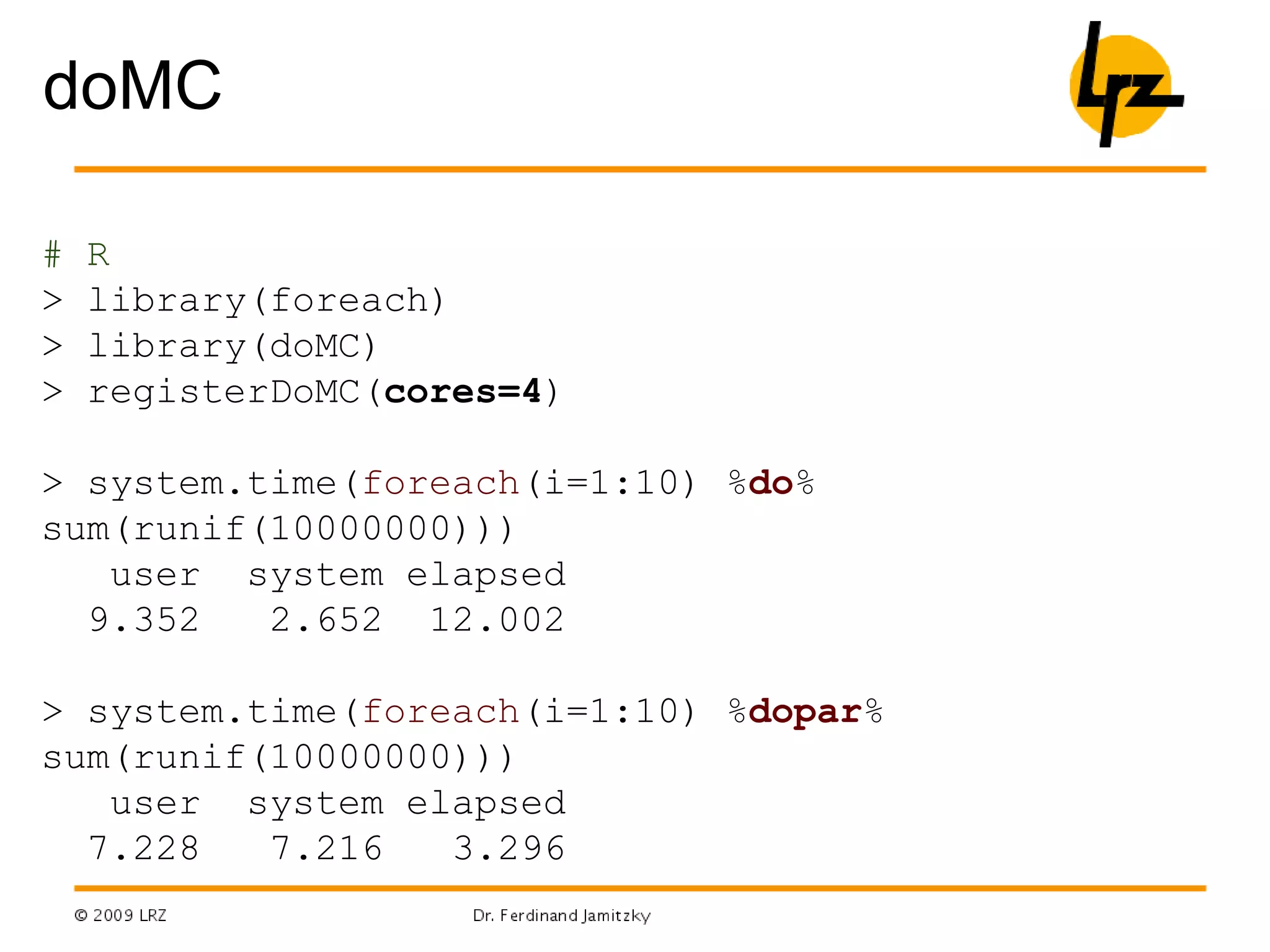
- Parallel programming, packages like foreach and doMC/doMPI/doSNOW allow parallel execution on multicore CPUs or computer clusters using MPI.



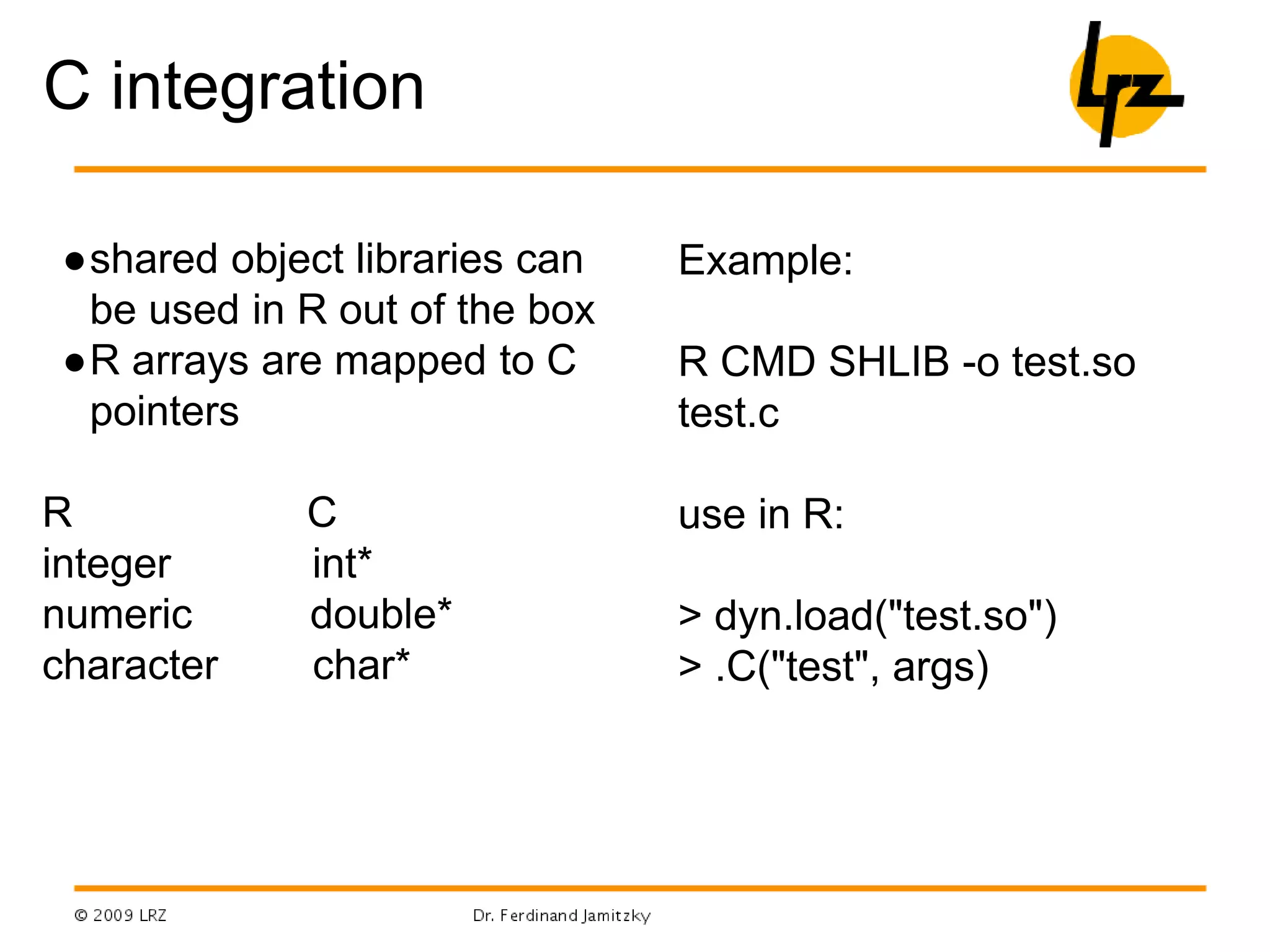
![C example
test.c:
void test(double* x, double* v, int *np, double *dt) {
int i;
double dx,dv;
for(i=0; i< *np; i++) {
dx=v[i]* *dt; dv=-x[i]* *dt;
x[i]+=dx; v[i]+=dv;
}
}
in R:
.C("test",as.numeric(runif(10)),as.numeric(runif(10)),as.integer(
10), as.numeric(0.01))](https://image.slidesharecdn.com/lrzkurserassuperglue-150320052836-conversion-gate01/75/Lrz-kurse-r-as-superglue-5-2048.jpg)


![Load and run
Load dynamic libraries
> dyn.load("mysub_host.so"), dyn.load("mysub_cuda.so"); np=1000000
Benchmark
> system.time(str(.Fortran("mysub_host",x=numeric(np),v=numeric(np),nstep=as.integer(1000))))
total energy: 666667.6633012500
total energy: 667334.6641391169
List of 3
$ x : num [1:1000000] -3.01e-07 -6.03e-07 -9.04e-07 -1.21e-06 -1.51e-06 ...
$ v : num [1:1000000] 1.38e-06 2.76e-06 4.15e-06 5.53e-06 6.91e-06 ...
$ nstep: int 1000
user system elapsed
26.901 0.000 26.900
> system.time(str(.Fortran("mysub_cuda",x=numeric(np),v=numeric(np),nstep=as.integer(1000))))
total energy: 666667.6633012500
total energy: 667334.6641391169
List of 3
$ x : num [1:1000000] -3.01e-07 -6.03e-07 -9.04e-07 -1.21e-06 -1.51e-06 ...
$ v : num [1:1000000] 1.38e-06 2.76e-06 4.15e-06 5.53e-06 6.91e-06 ...
$ nstep: int 1000
user system elapsed
0.829 0.000 0.830
Acceleration Factor:
> 26.9/0.83
[1] 32.40964](https://image.slidesharecdn.com/lrzkurserassuperglue-150320052836-conversion-gate01/75/Lrz-kurse-r-as-superglue-9-2048.jpg)