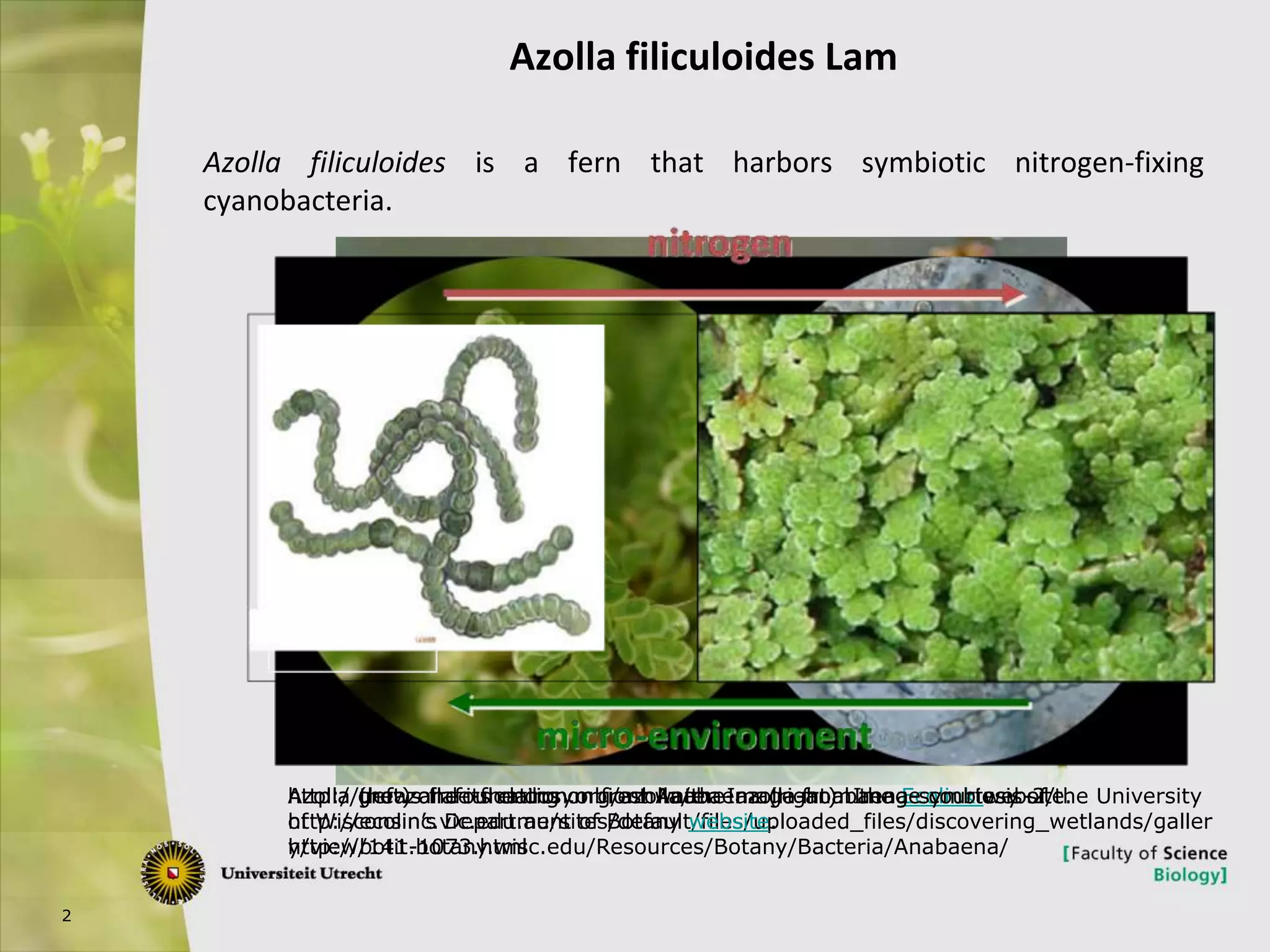
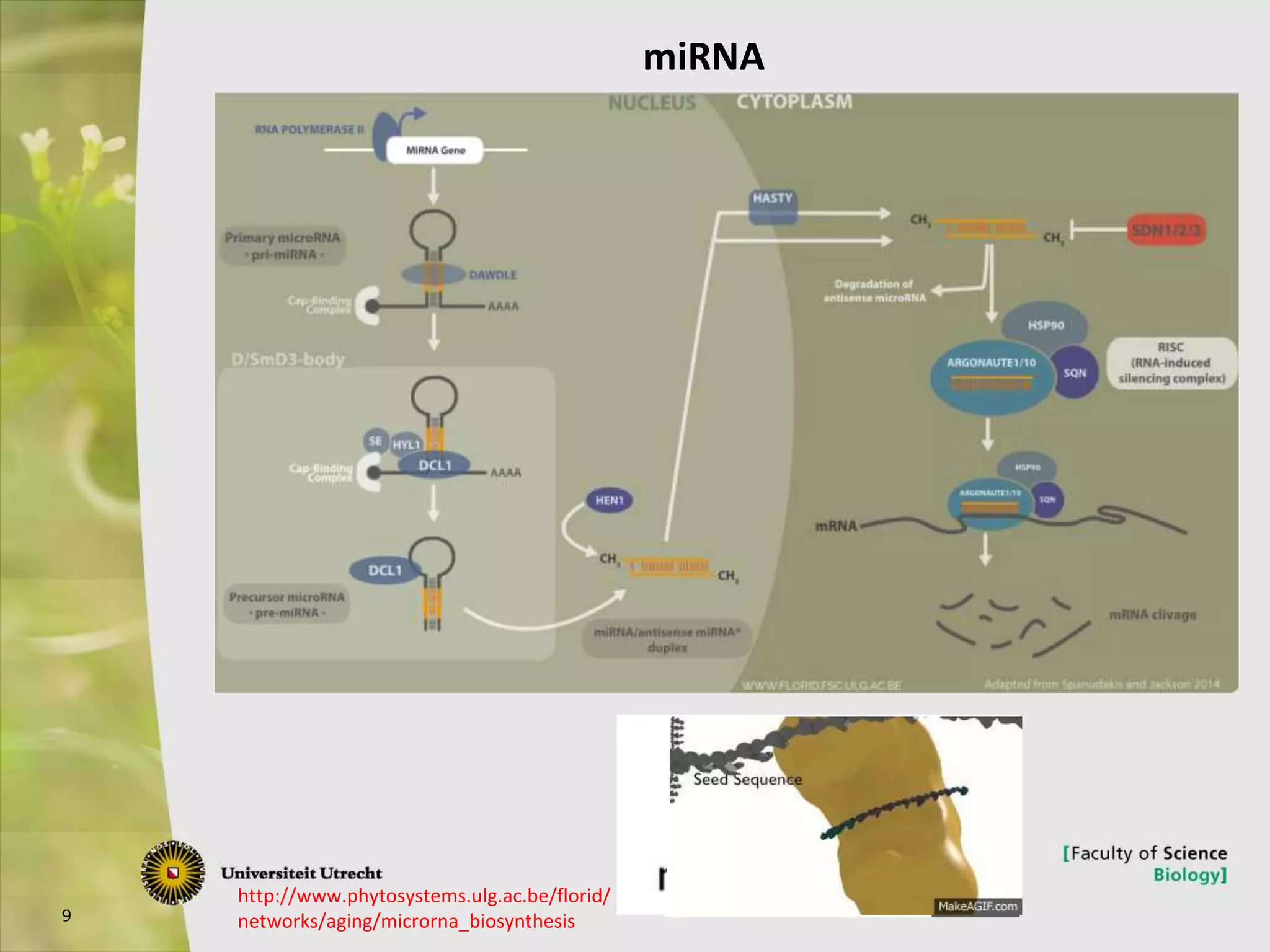
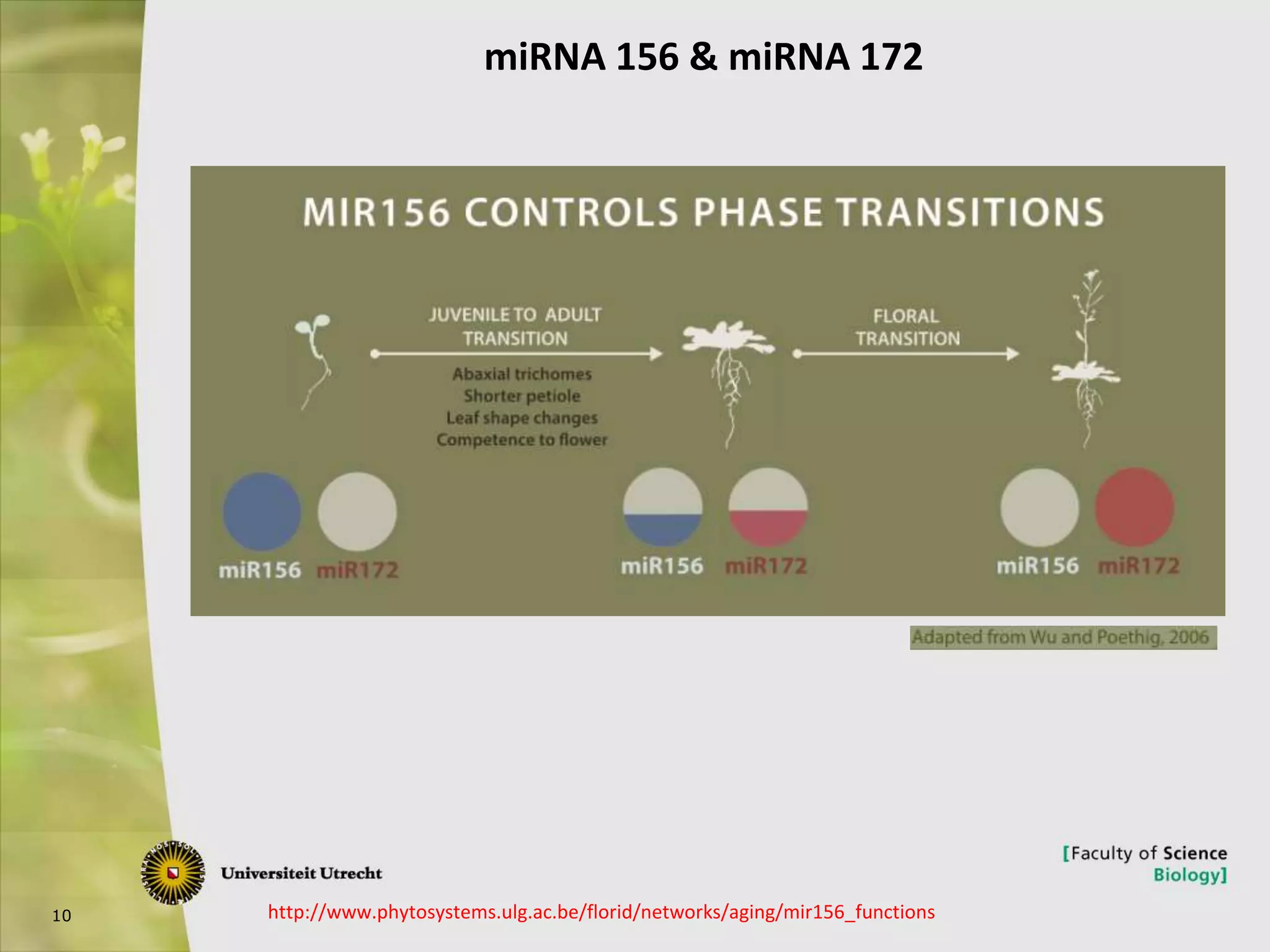
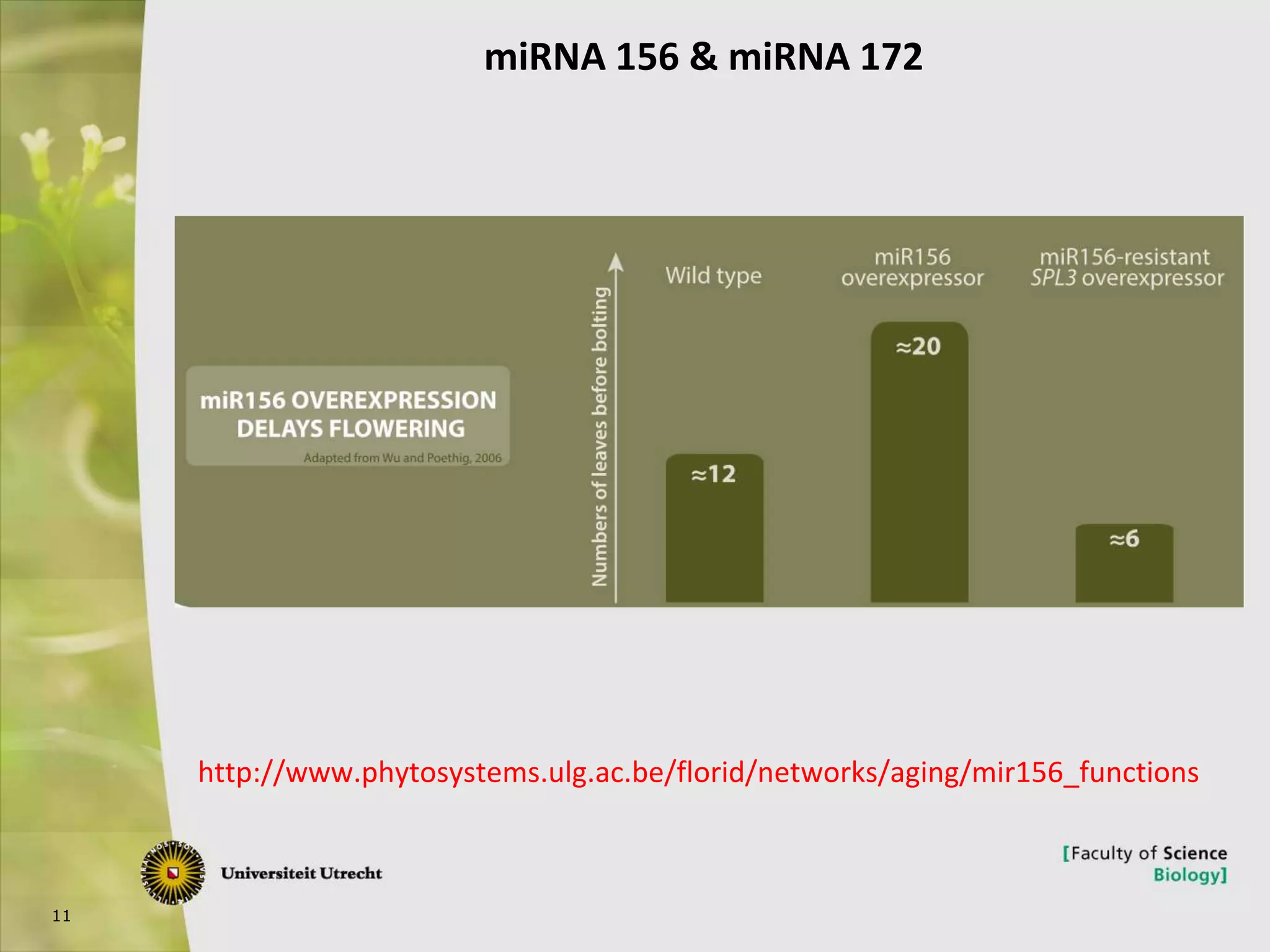
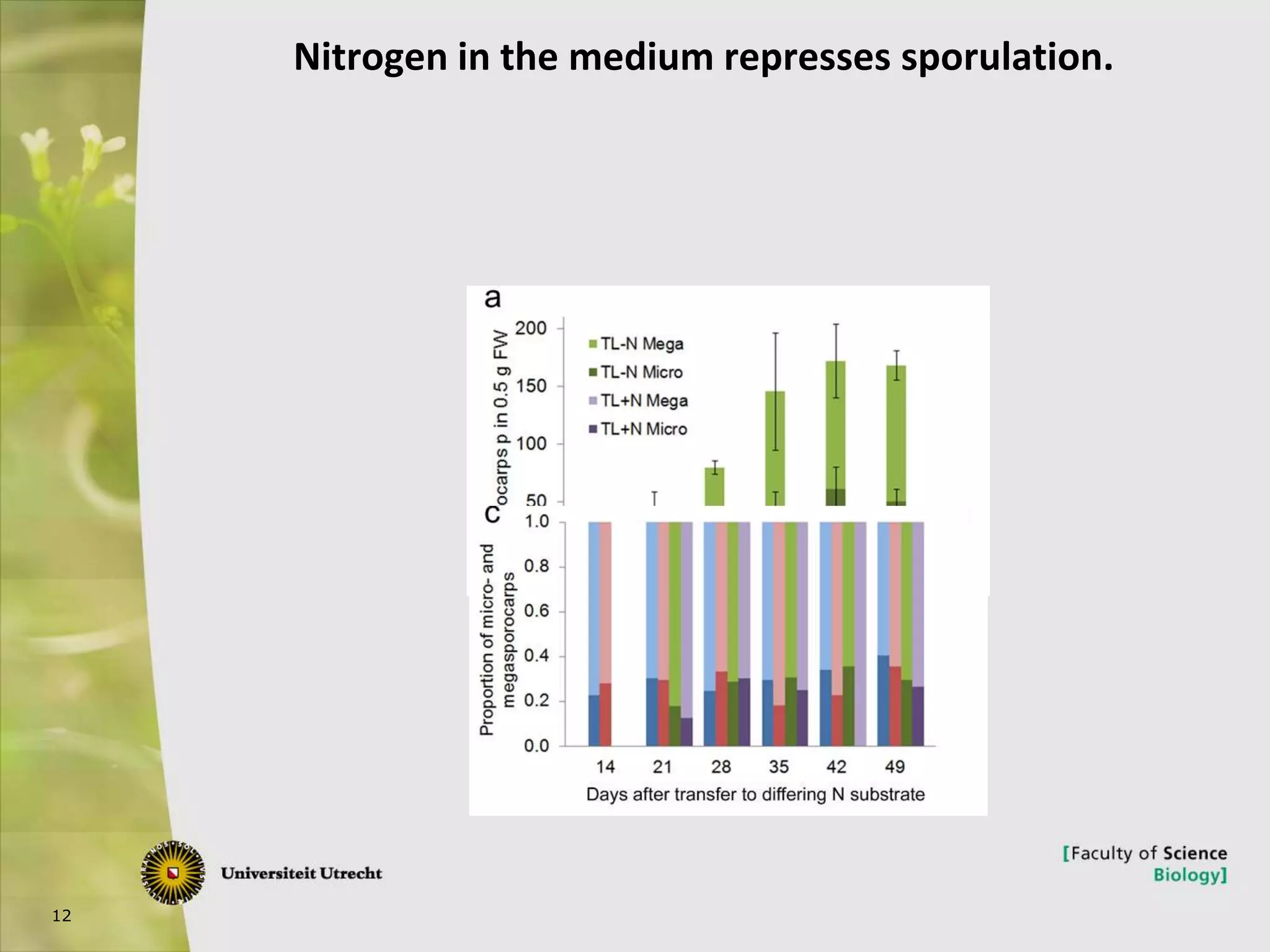
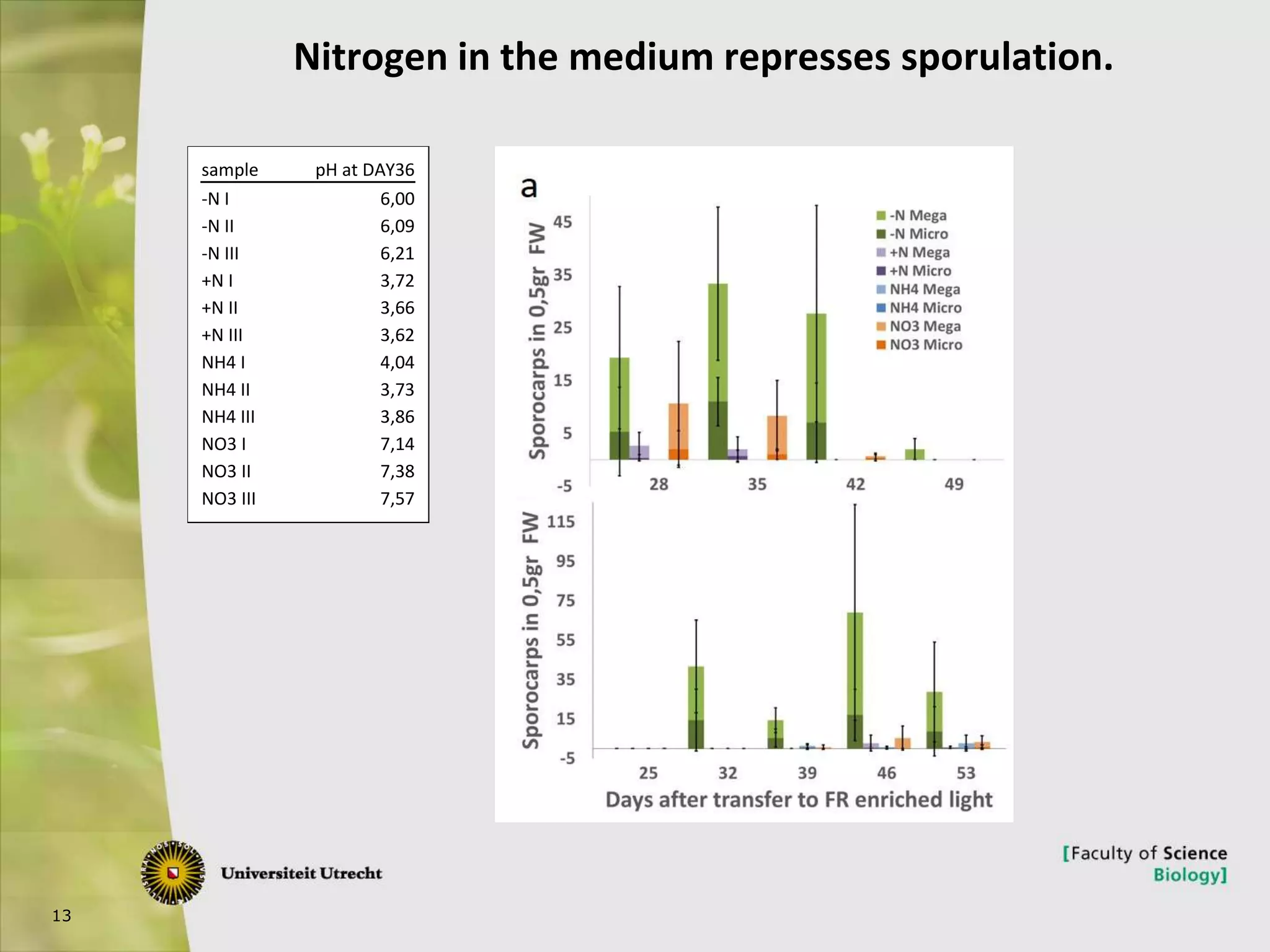
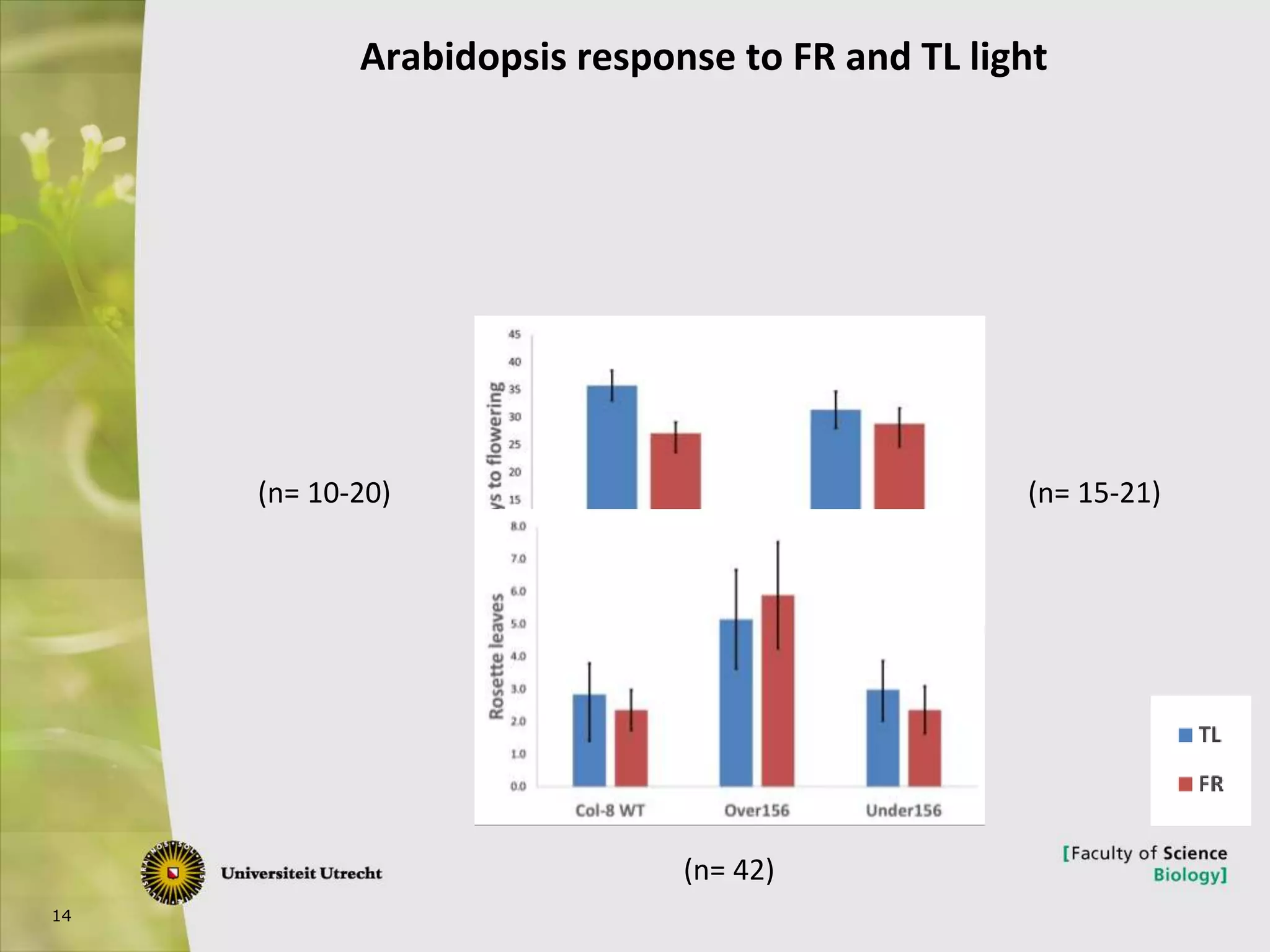
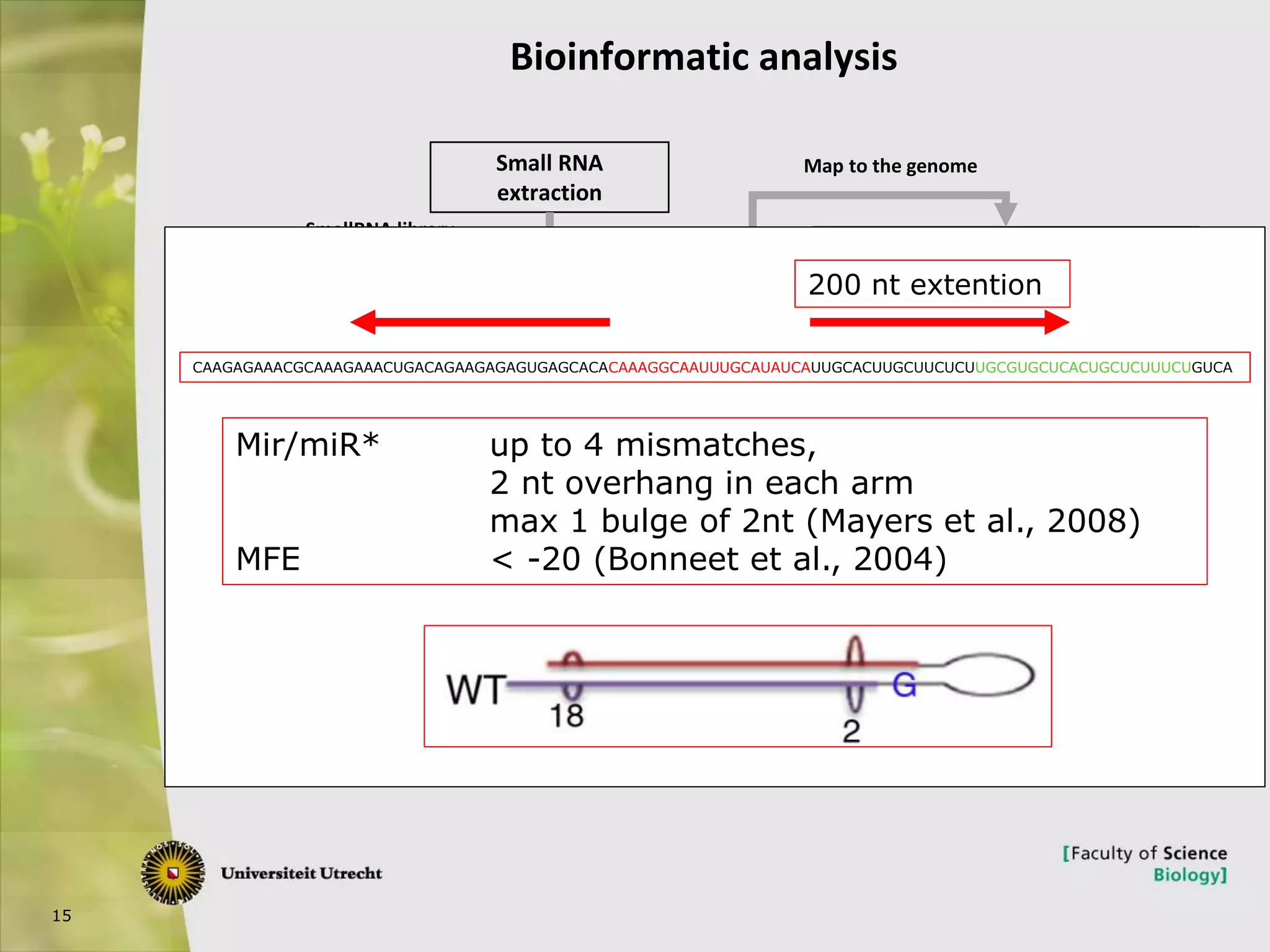
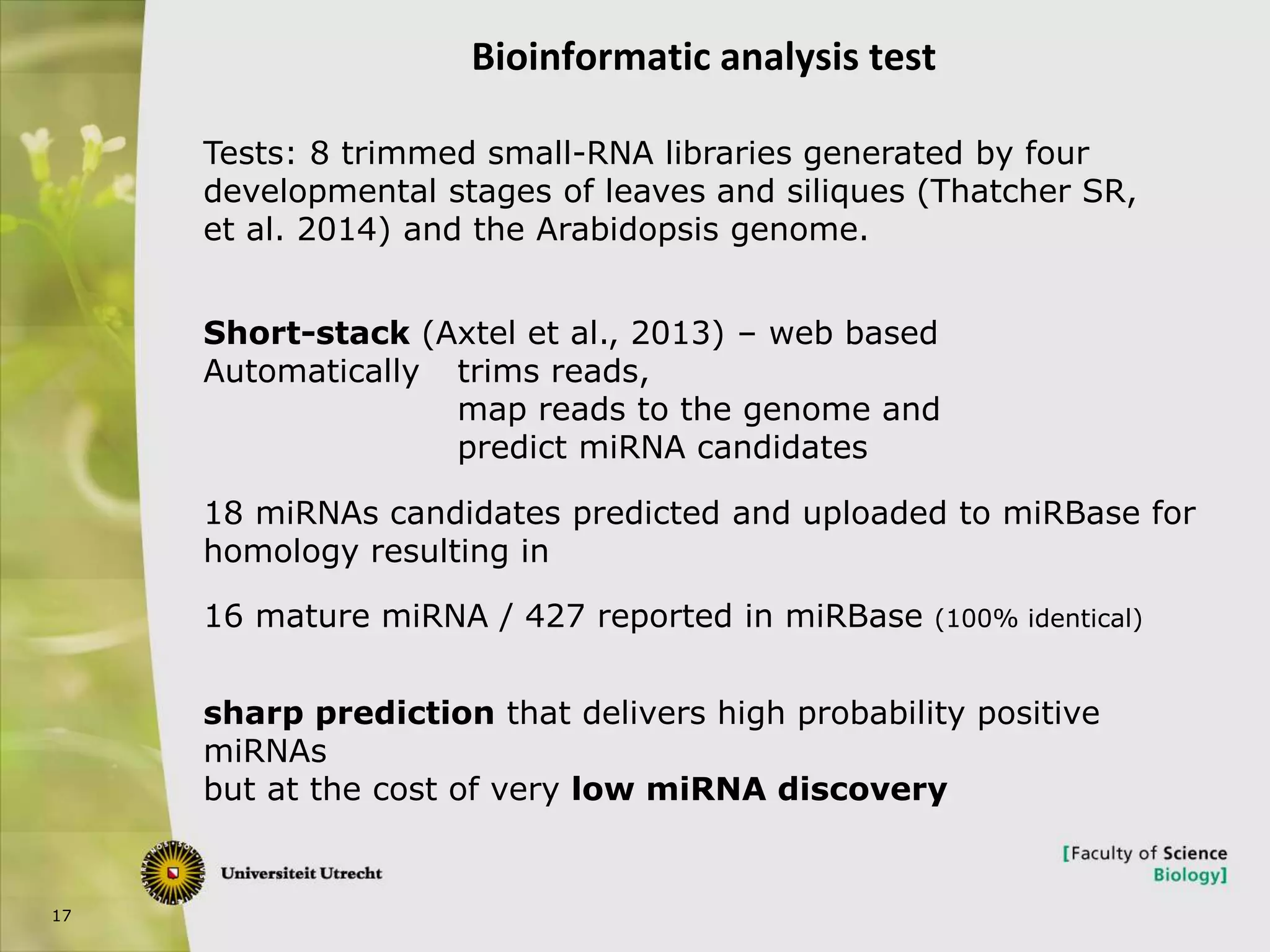
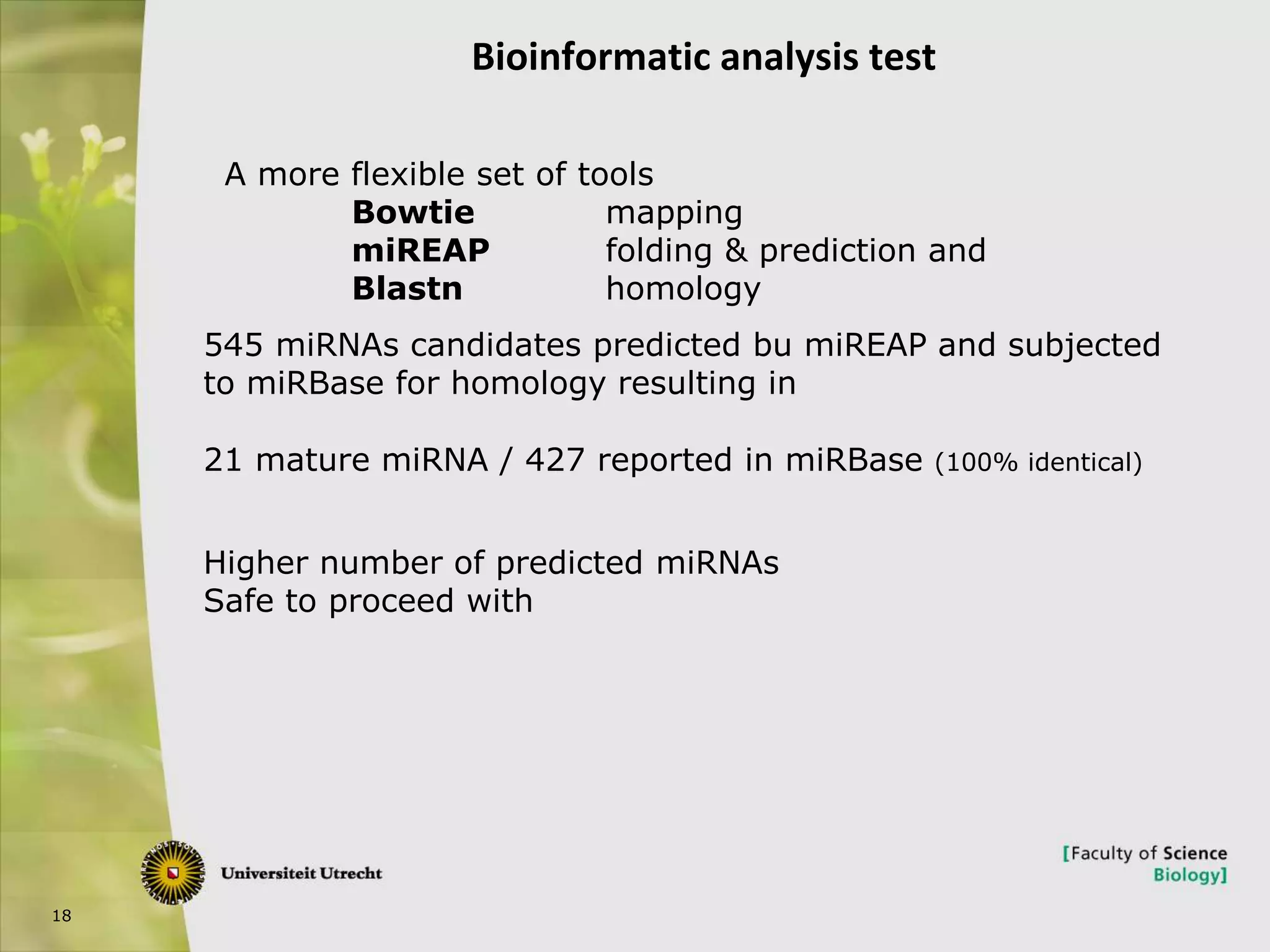
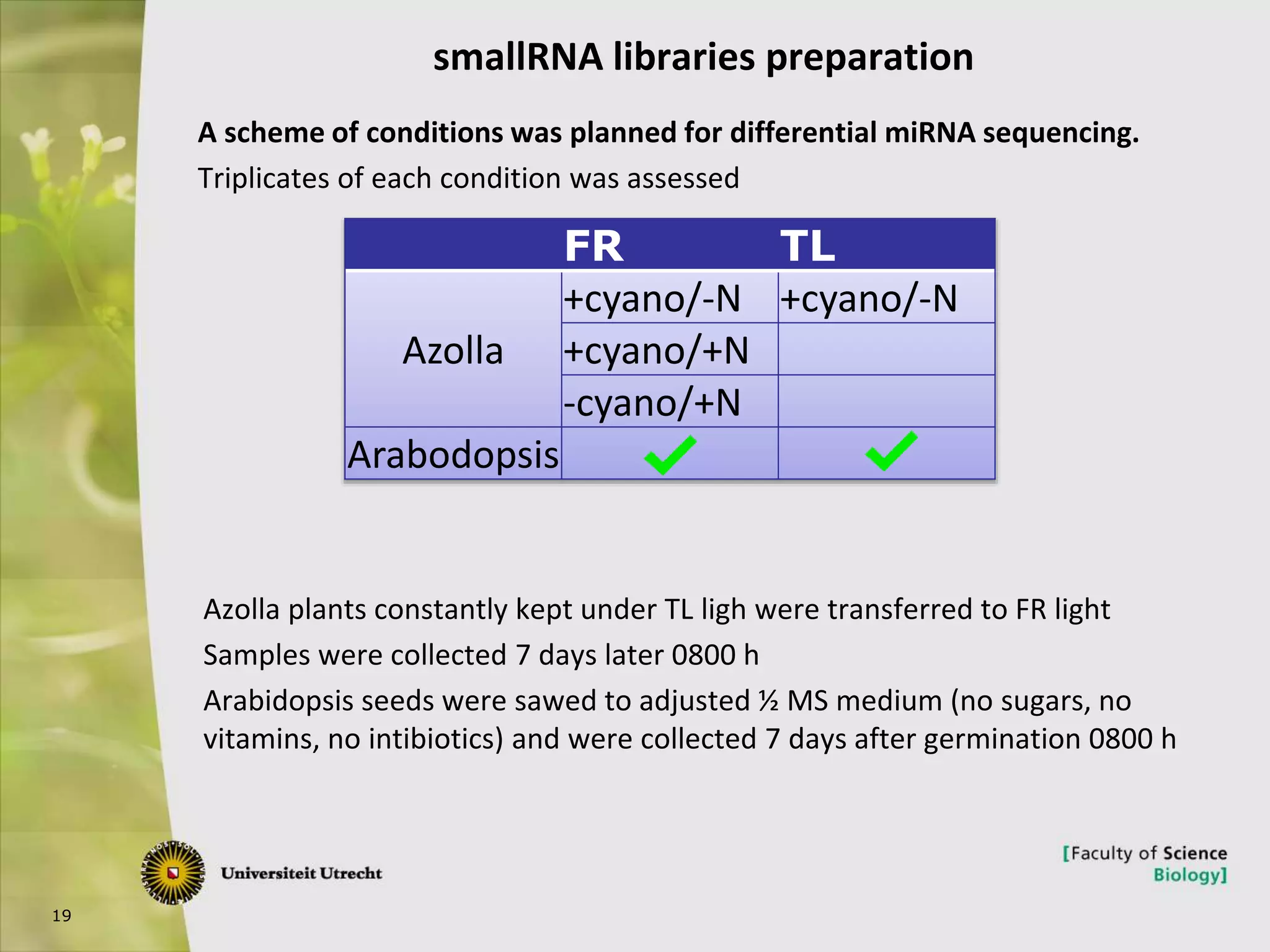
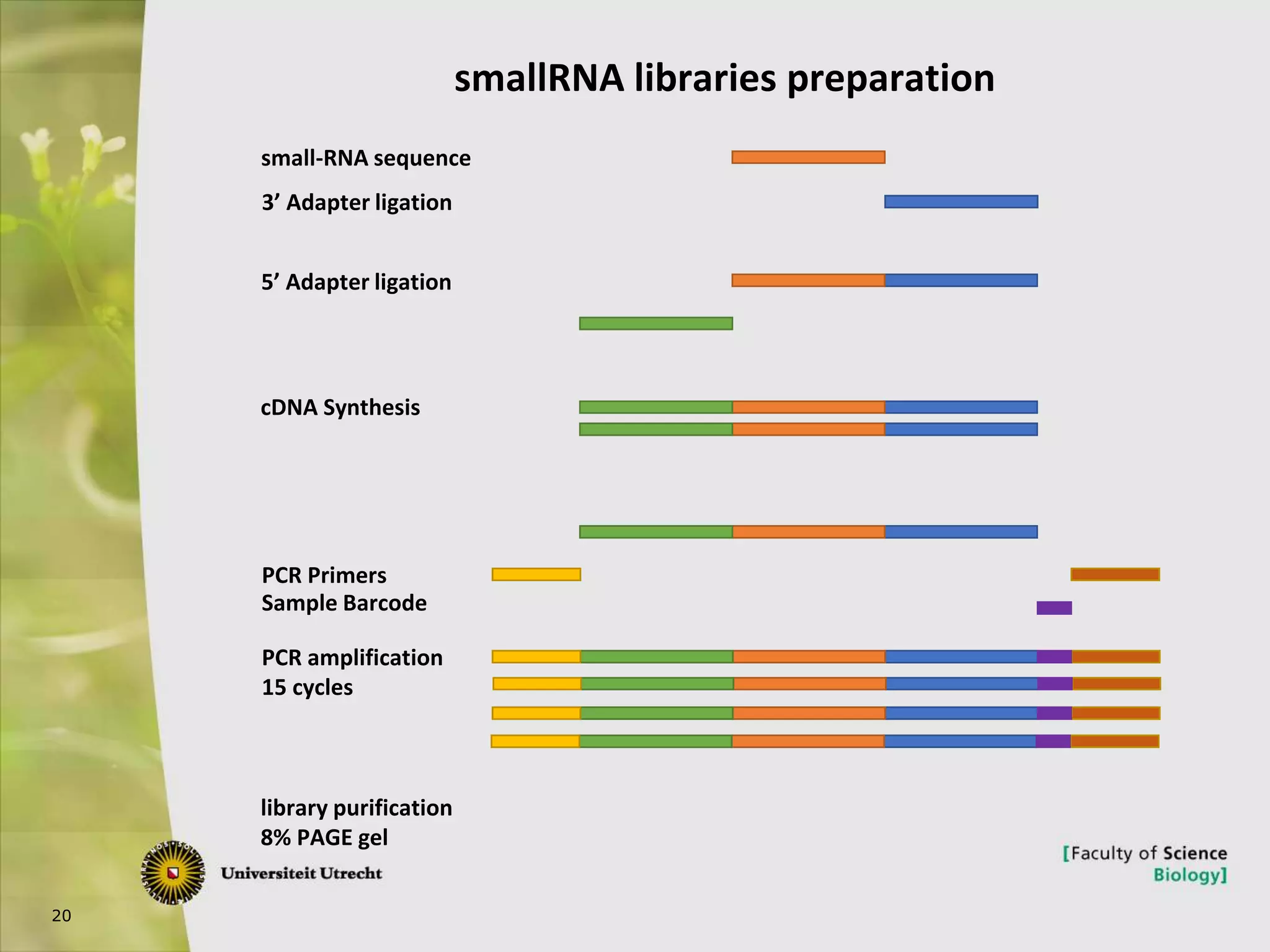
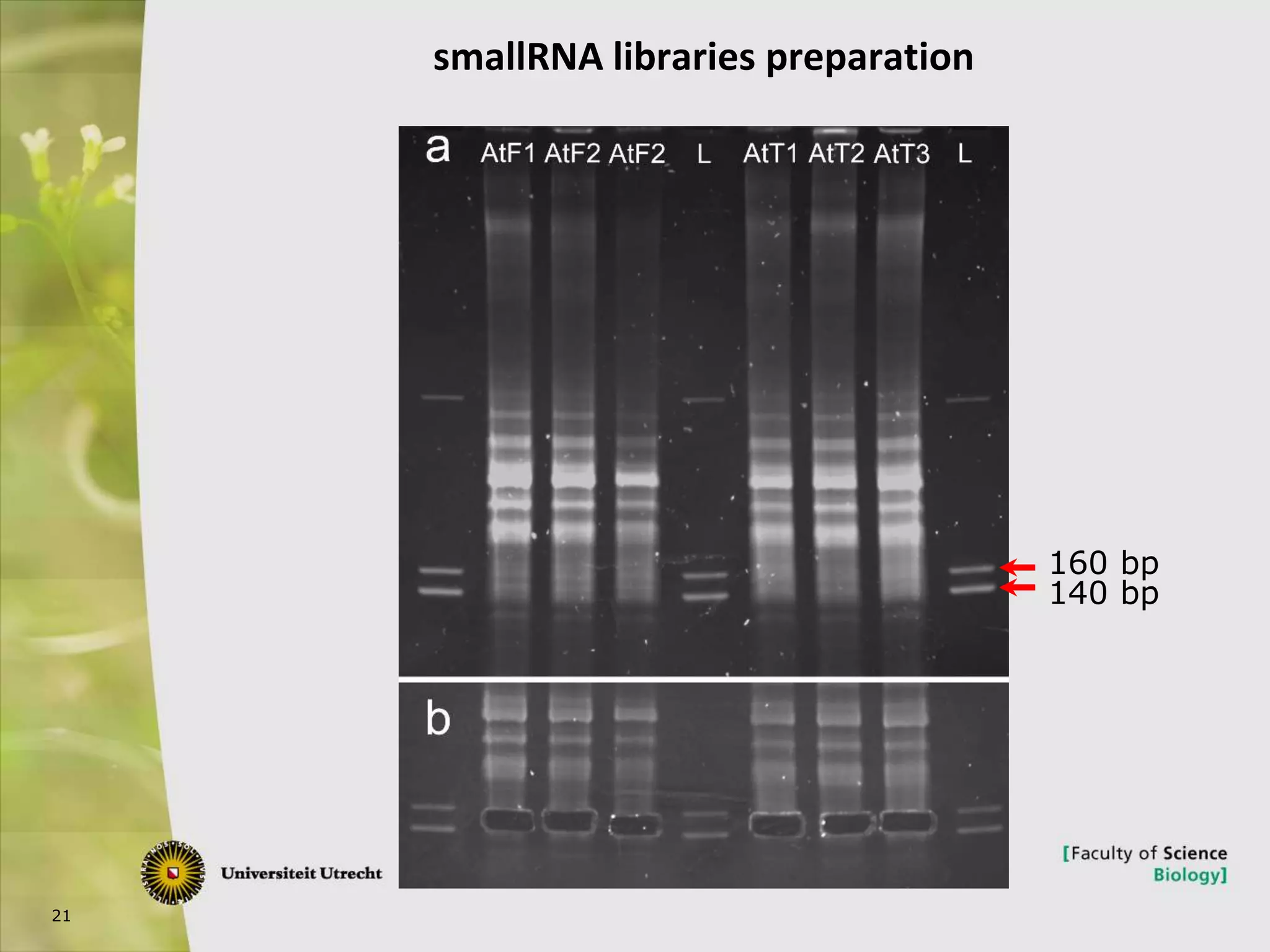
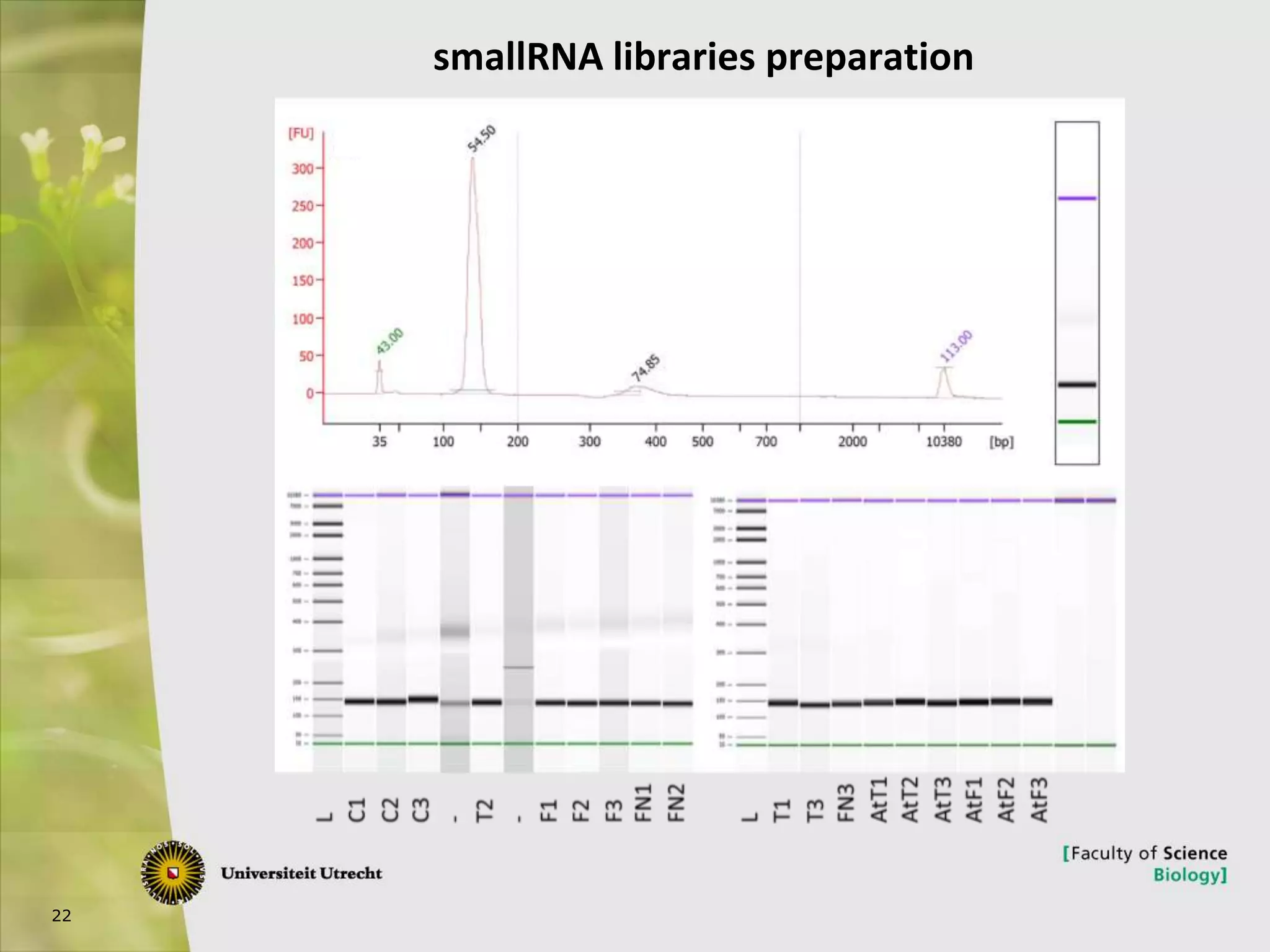
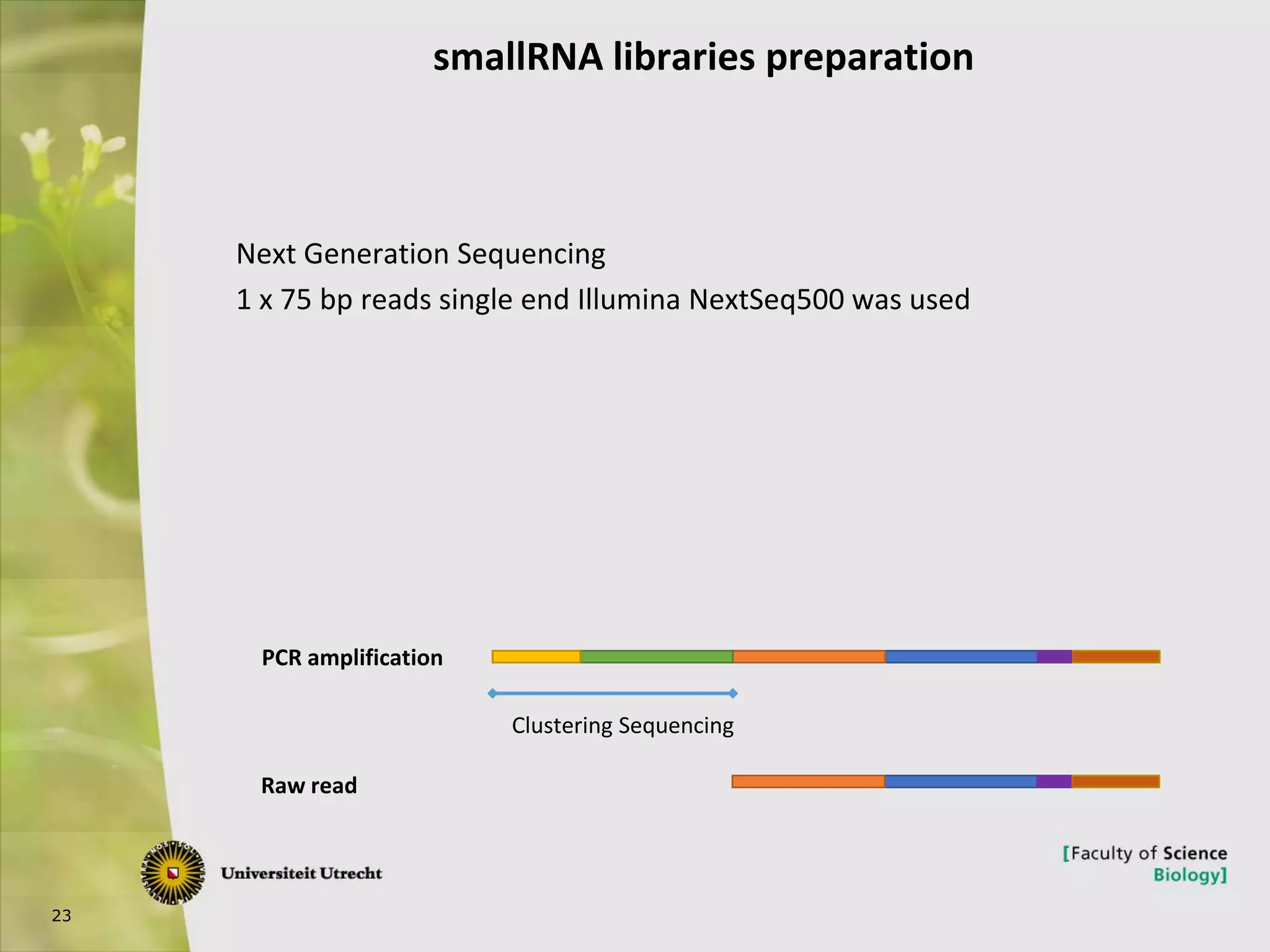
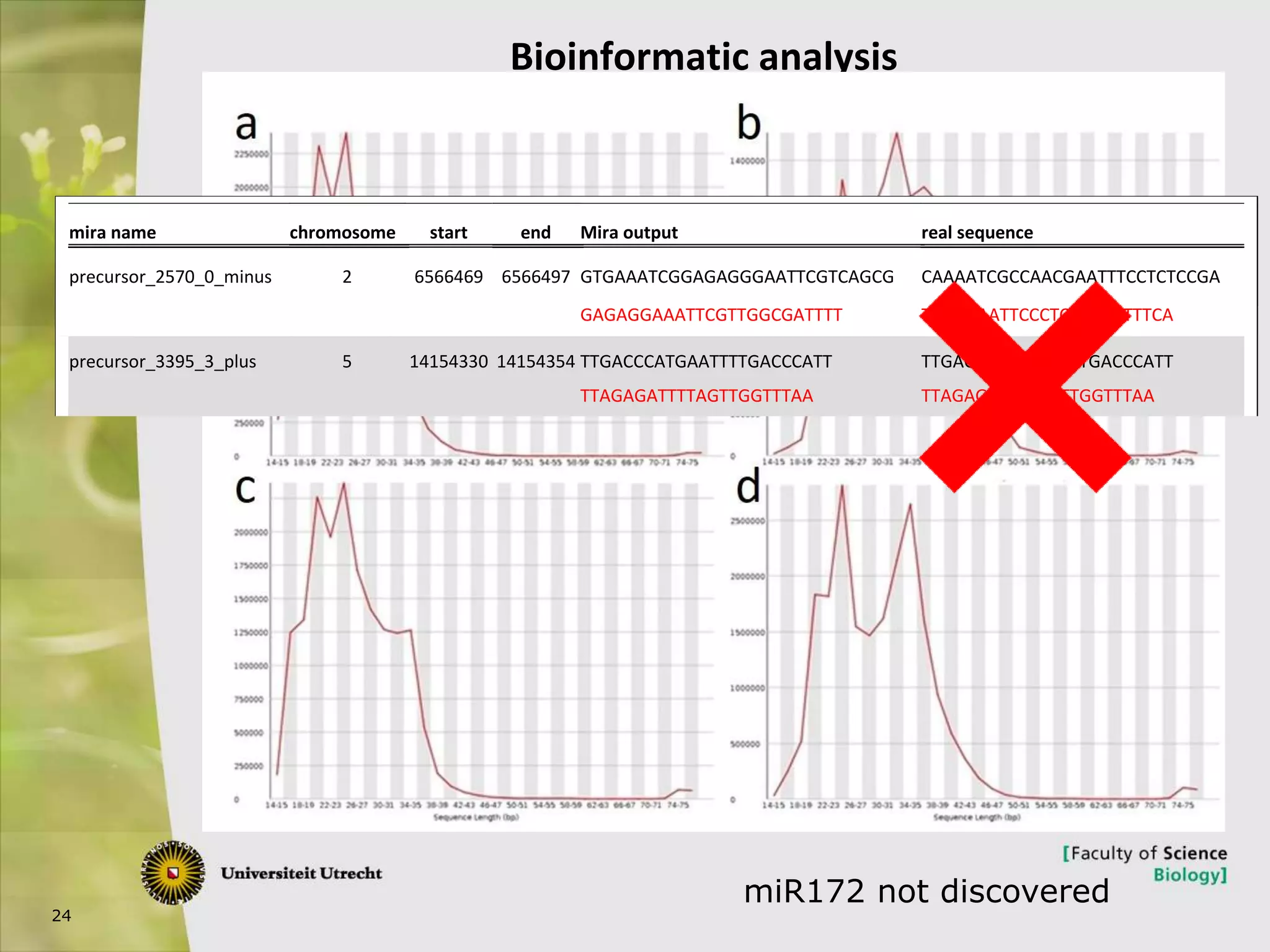
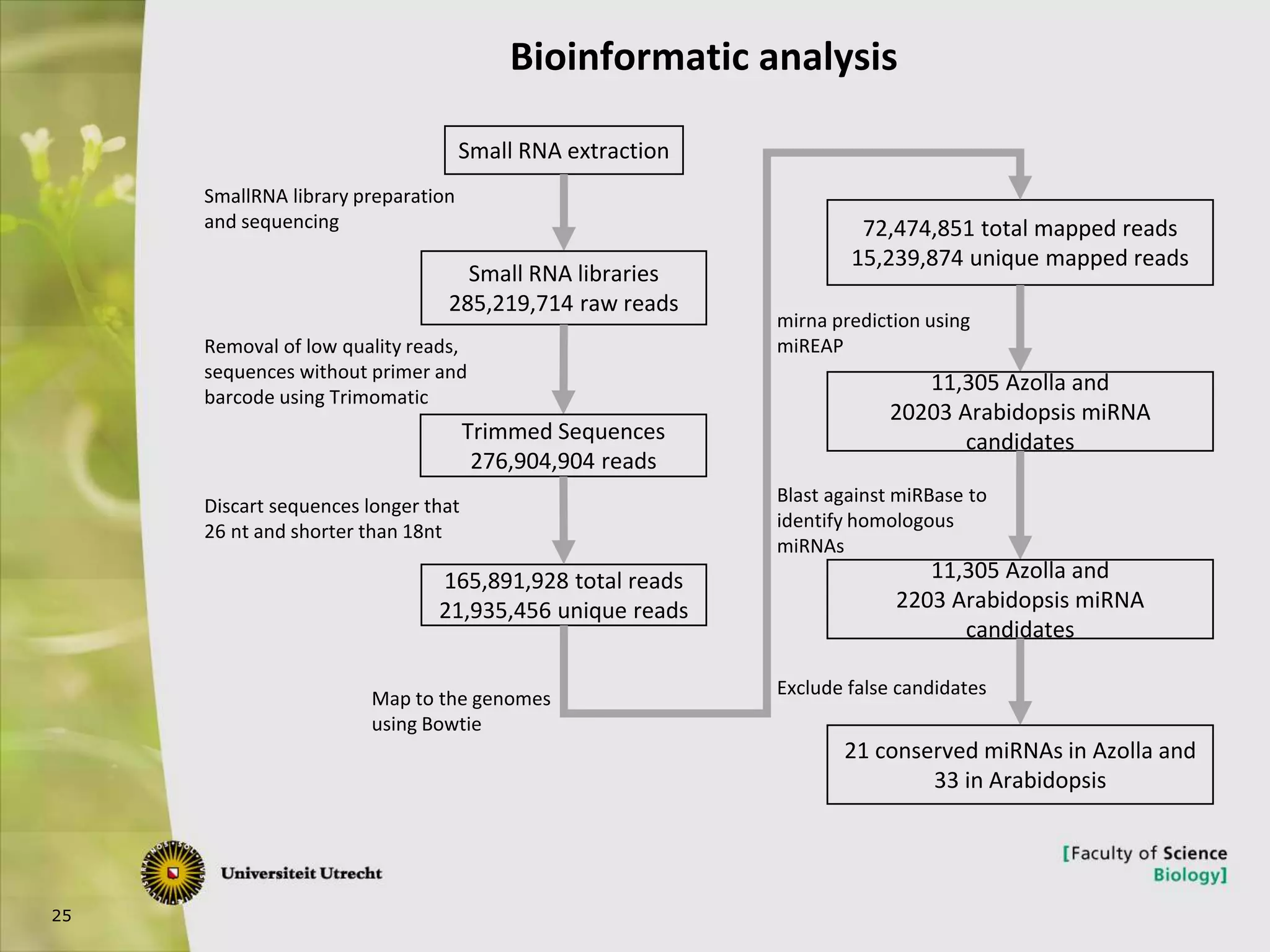
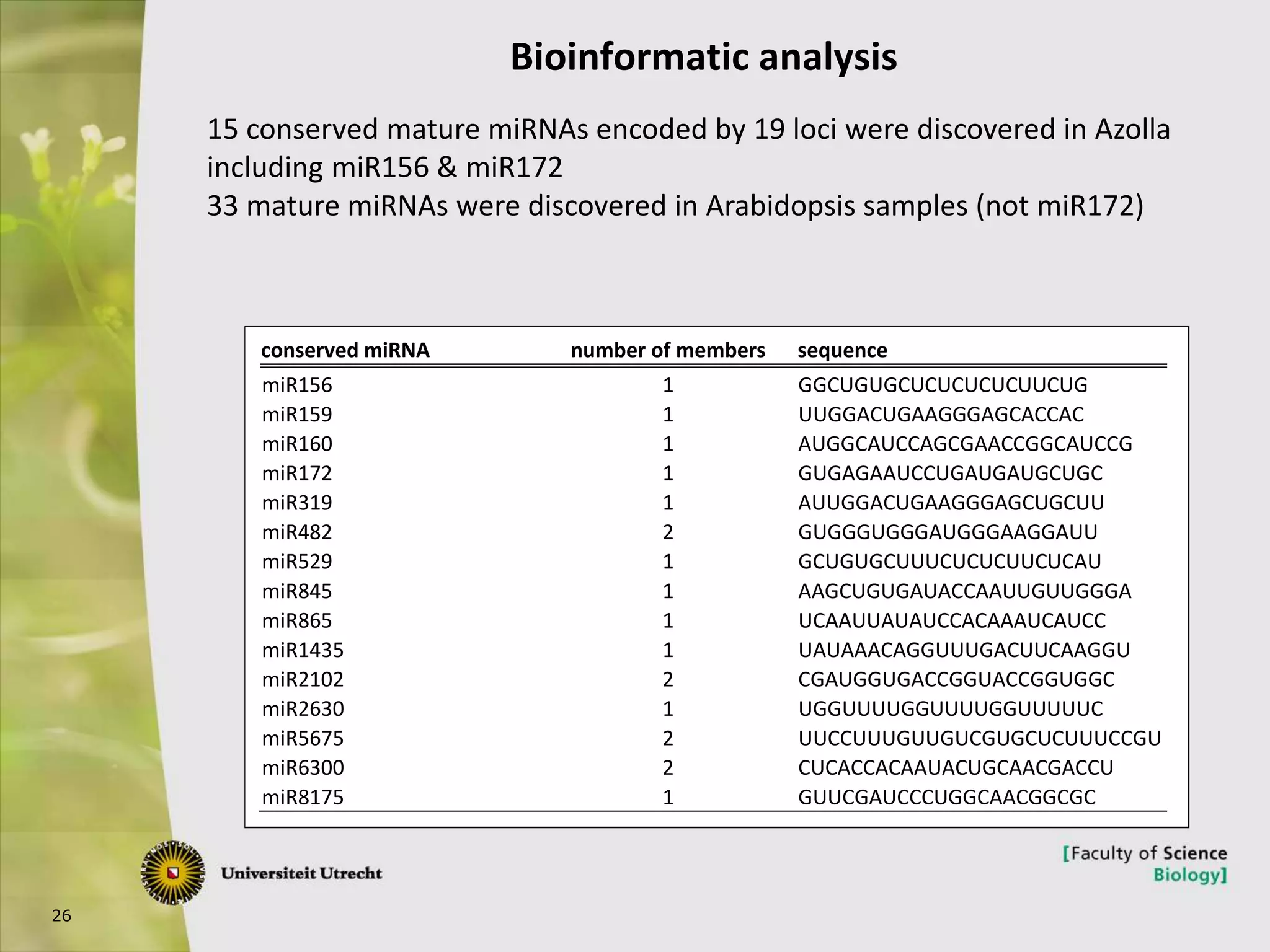
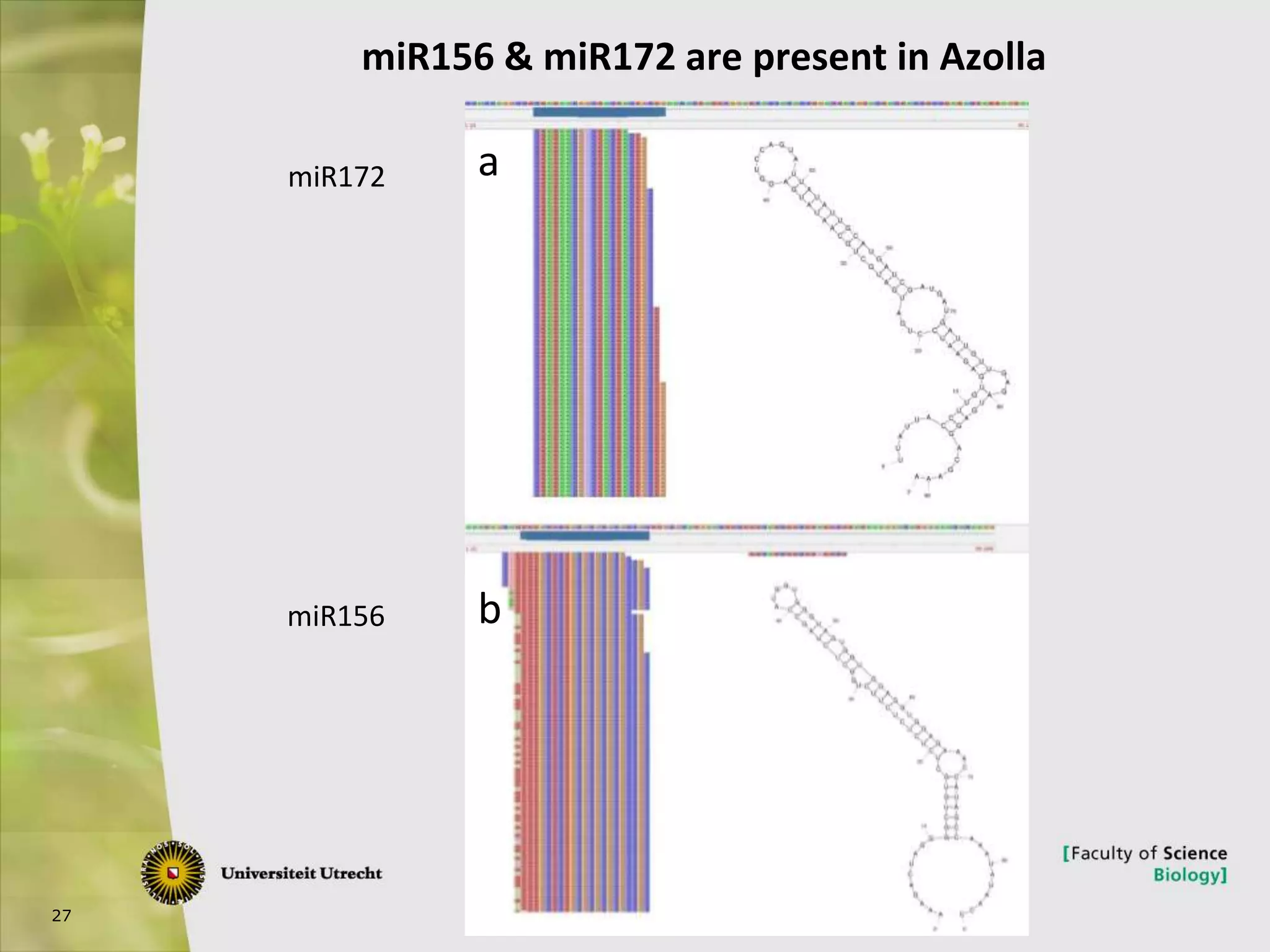
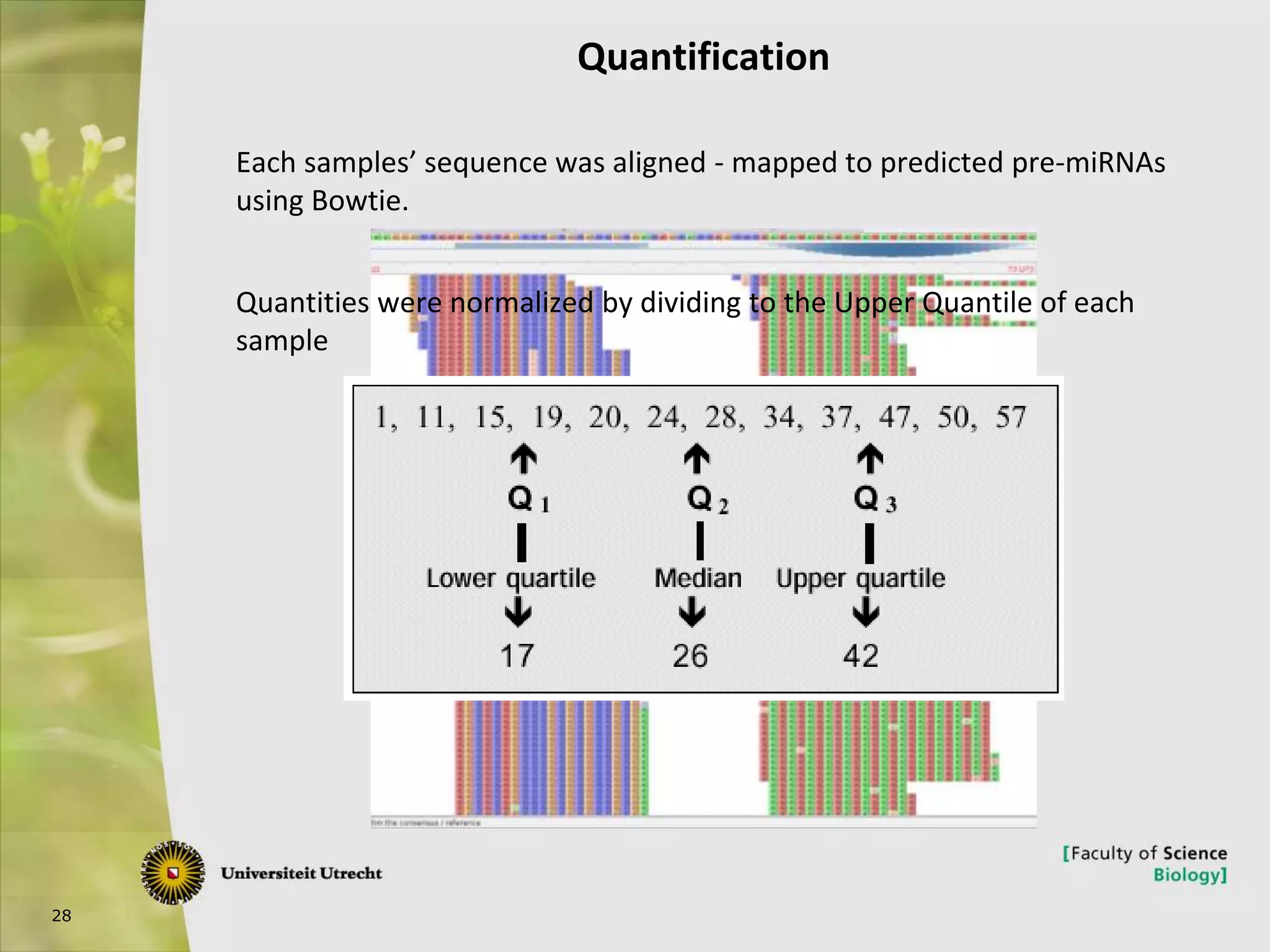
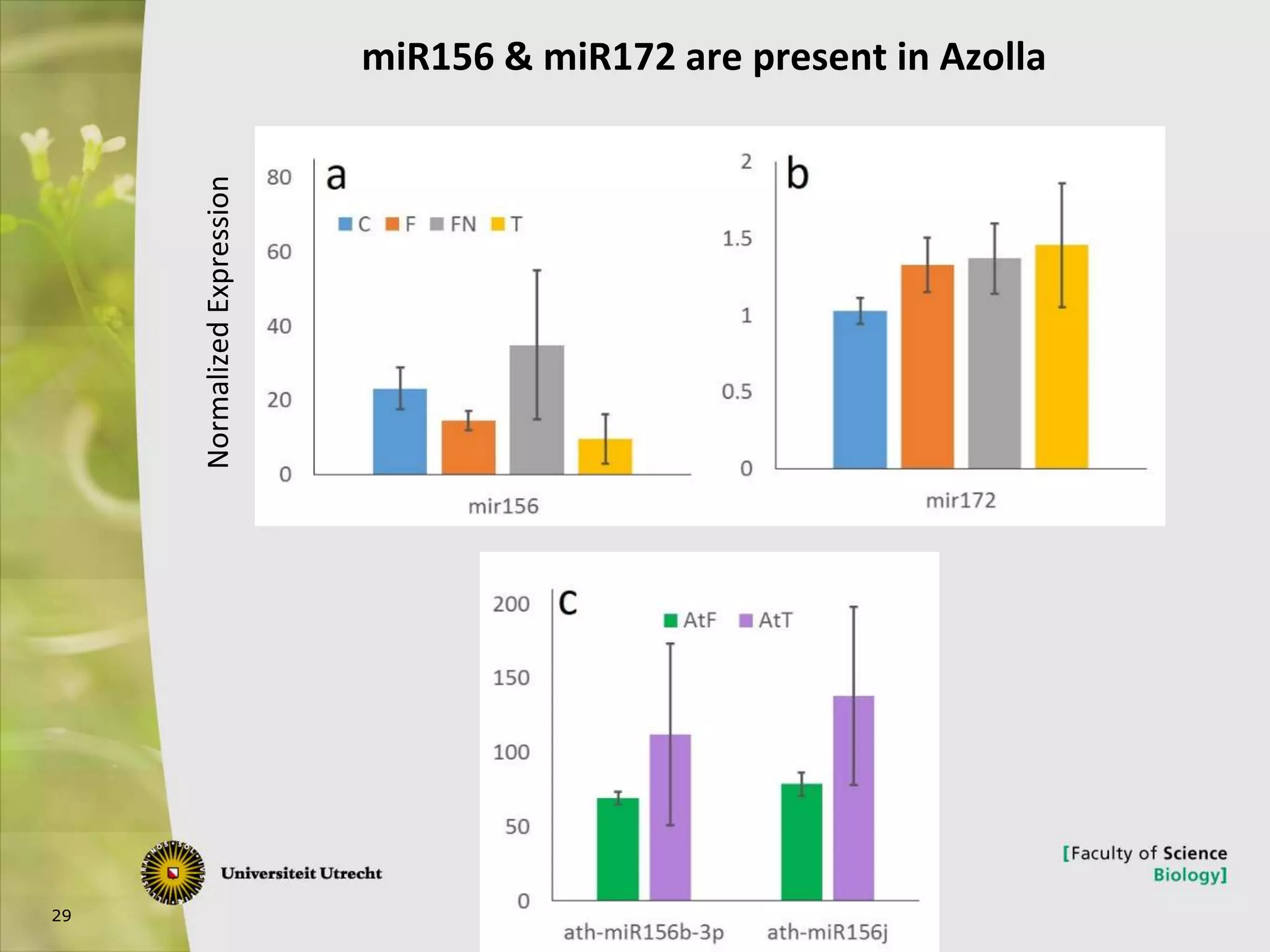
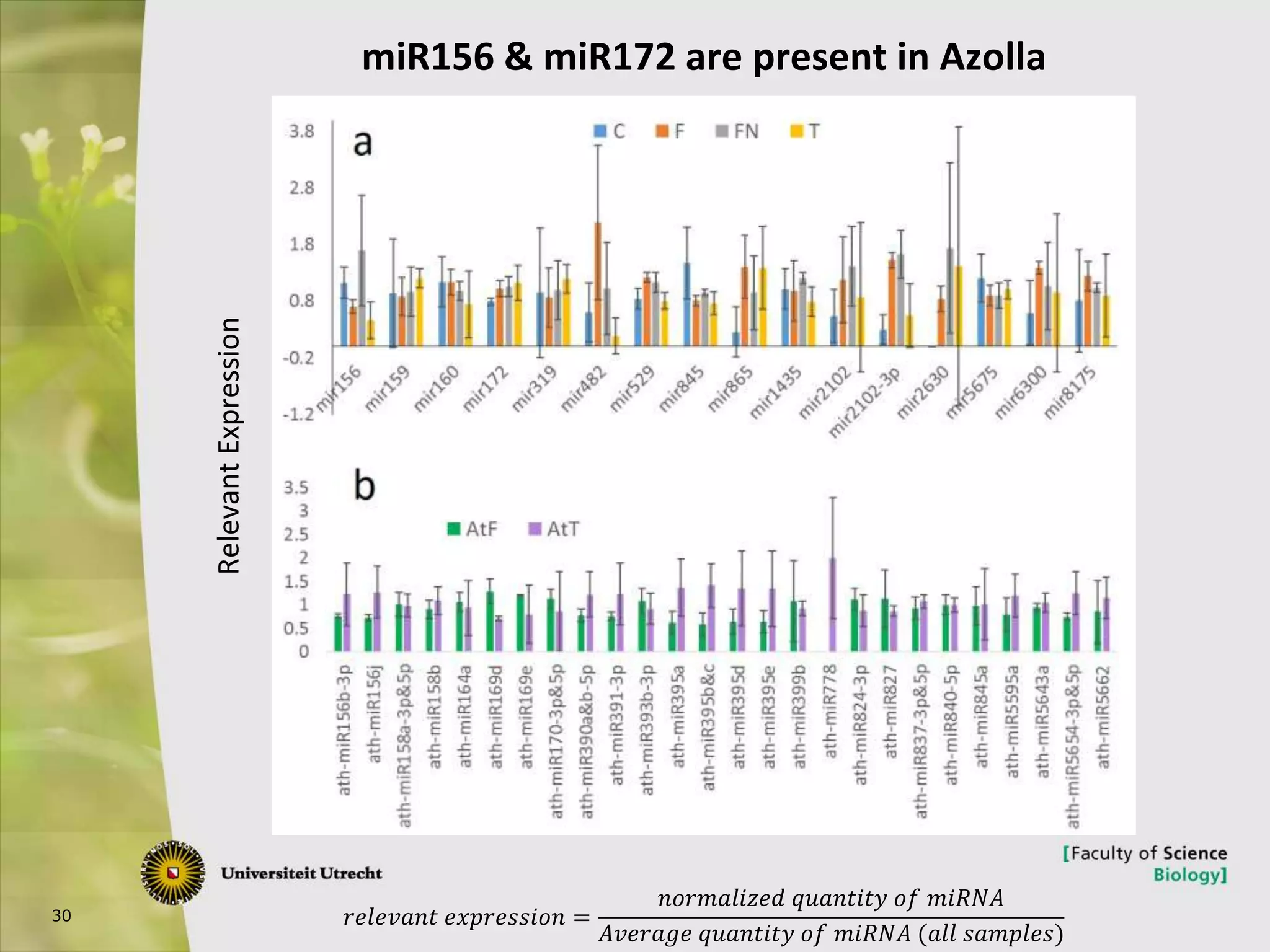
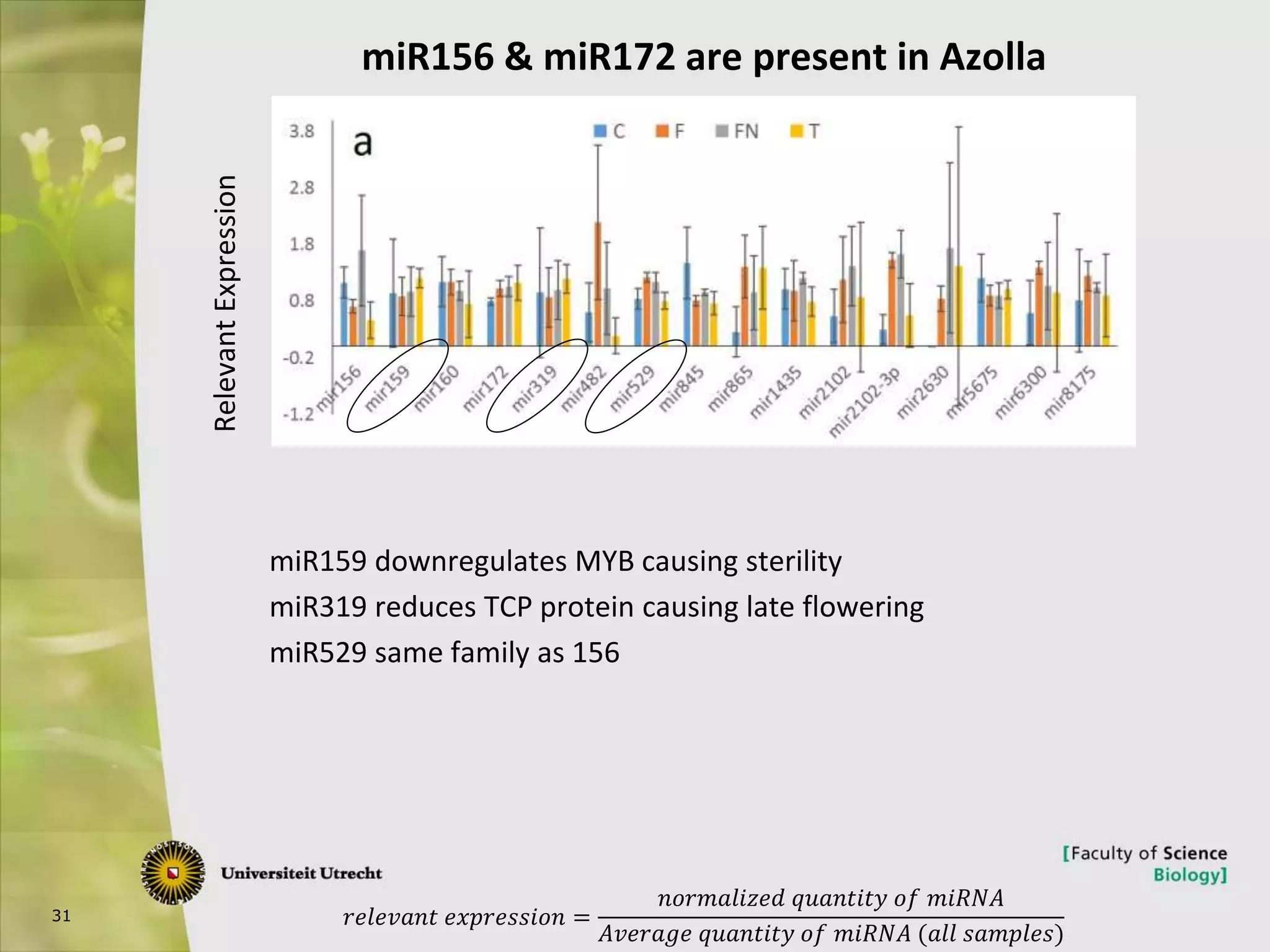
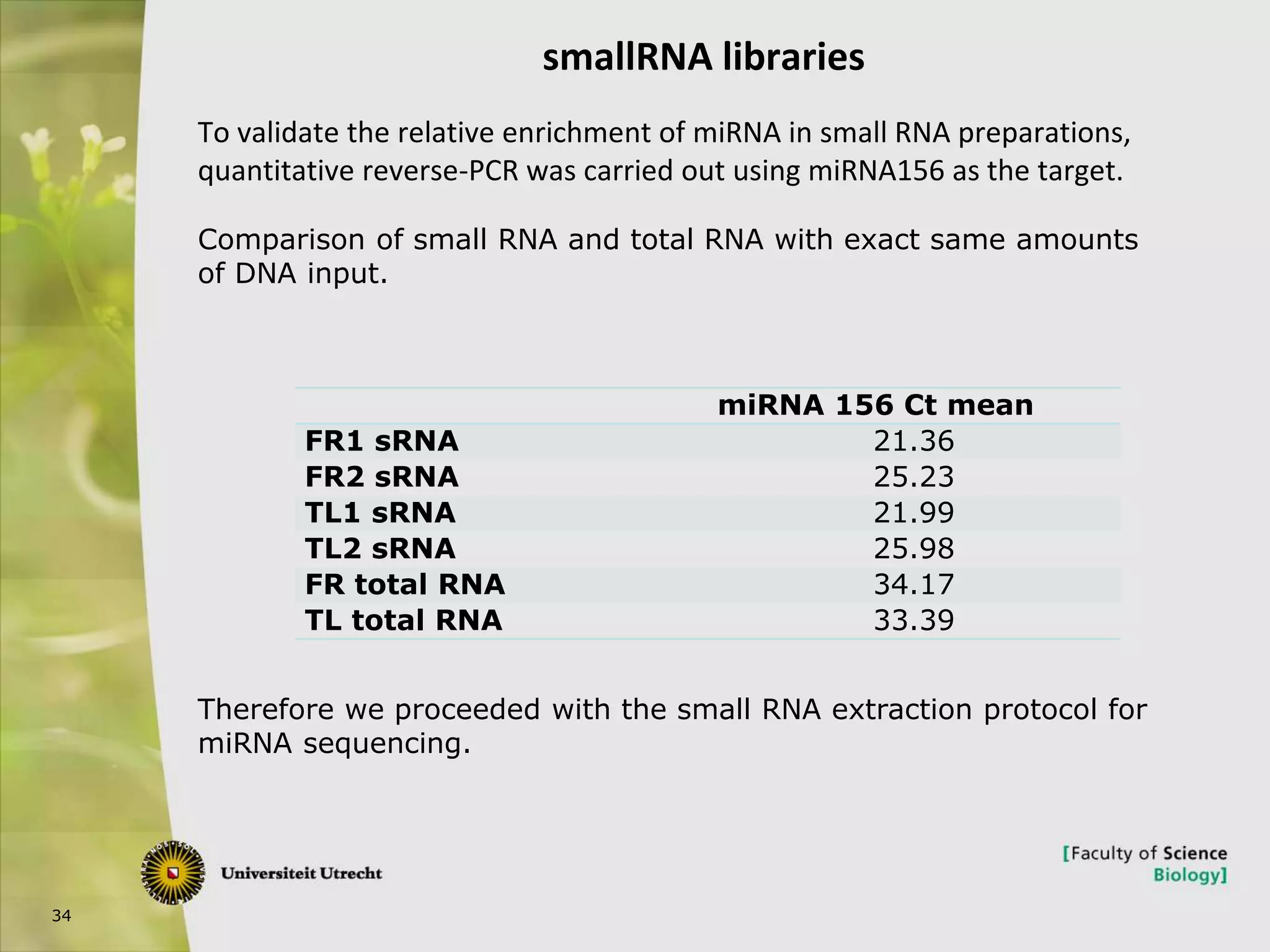
The document describes research into controlling the reproductive cycle of Azolla filiculoides, a small aquatic fern. Through next-generation sequencing of small RNA libraries from Azolla under different conditions, 15 conserved microRNAs were discovered, including miR156 and miR172. While miR172 was not found, miR156 and miR172 are known to regulate flowering time in plants and their presence in Azolla suggests they may play a role in controlling sporulation. Quantification of small RNA sequencing data found that miR156 and miR172 are present in Azolla.