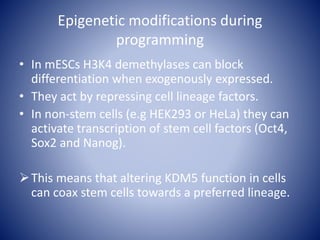
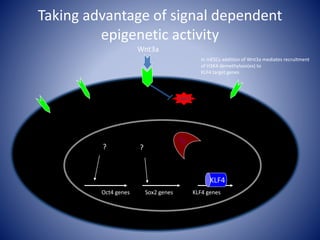
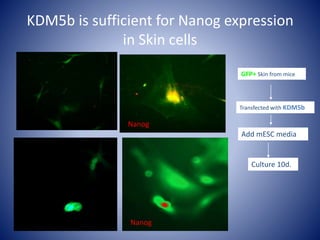
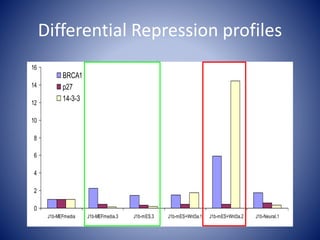
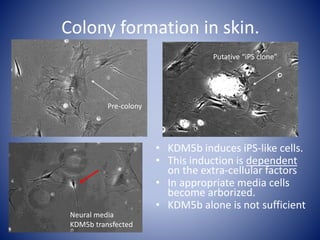
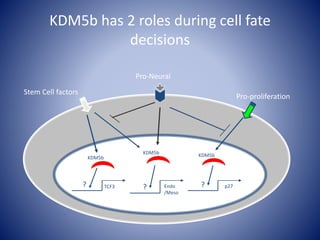
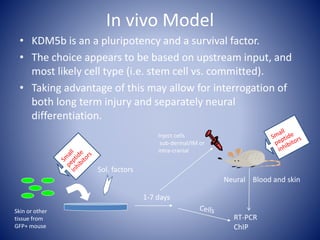
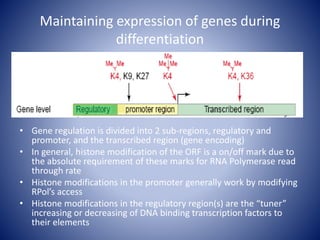
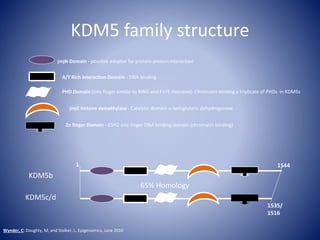
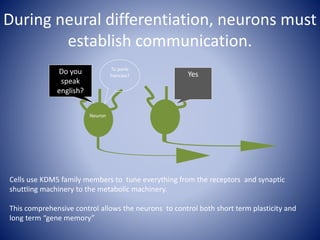
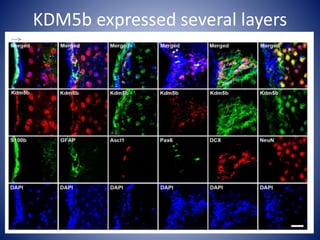
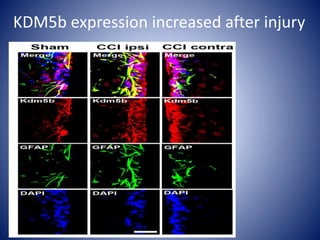
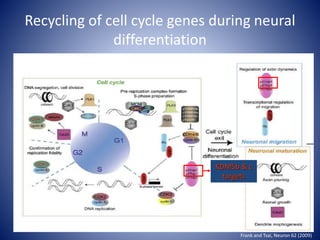
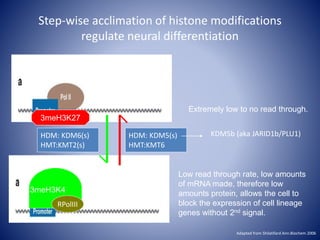
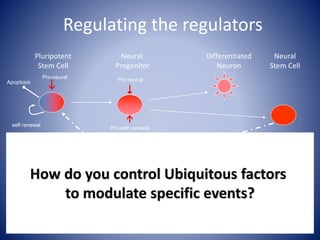
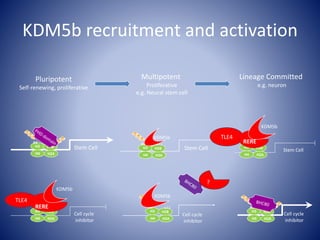
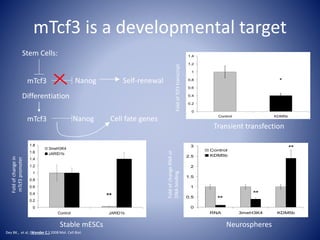
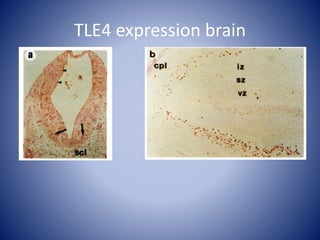
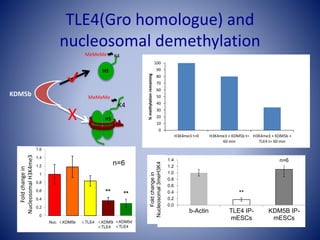
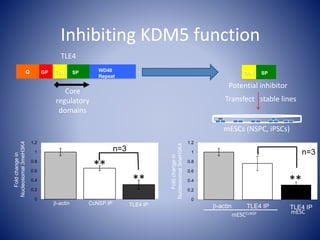
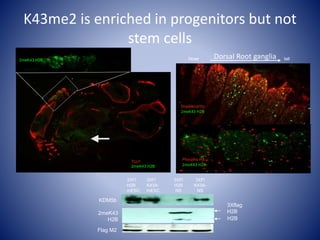
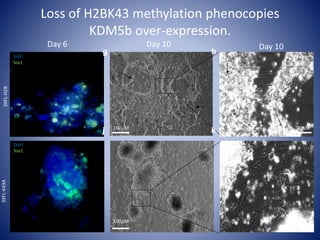
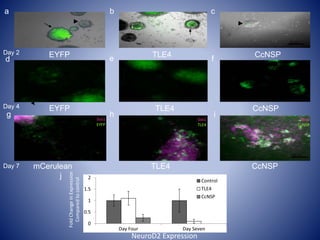
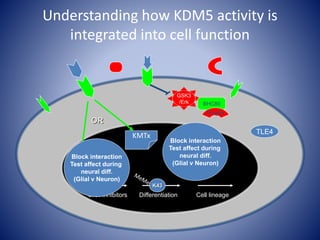
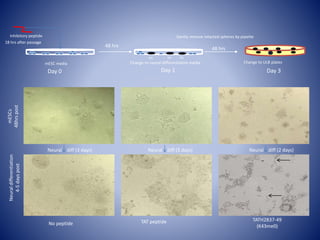
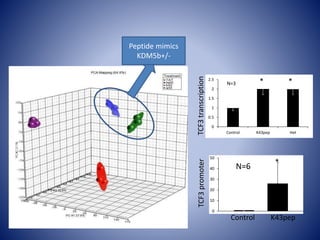
The document discusses the role of epigenetic enzymes, particularly the KDM5 family, as potential therapeutic targets for neurological diseases. It highlights how epigenetic regulation influences gene expression during neural differentiation and the importance of histone modifications in controlling neuron development and function. The text emphasizes the relevance of understanding these mechanisms for advancing stem cell therapies and addressing neurological disorders.



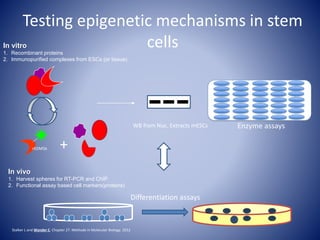
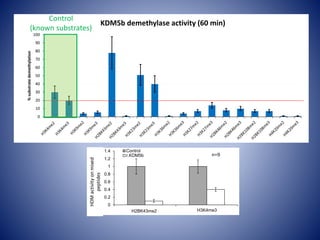
![0
10
20
30
40
50
60
70
80
90
100
0 30 60 90 120 150 180 210
%demethylatedsubstrate
Time [min]
H3K4me3
H2BK43me2
r.KDM5b removes methyl groups from K43
faster than K4](https://image.slidesharecdn.com/epigeneticdrugs-140817111058-phpapp02/85/KDM5-epigenetic-modifiers-as-a-focus-for-drug-discovery-22-320.jpg)
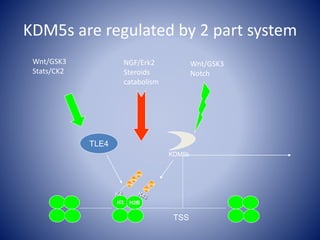
![Linking biochemistry to biological
properties-Future directions
1. Define relationship between KDM5 and
cell signaling (MS sequencing and
verification)
2. Define the cell biology that is altered by
modulation of this system (Post injury
NSCs and iPSCs [NSCs v skin])
3. What is the role of these proteins in
both injury and recovery (mouse
models)](https://image.slidesharecdn.com/epigeneticdrugs-140817111058-phpapp02/85/KDM5-epigenetic-modifiers-as-a-focus-for-drug-discovery-26-320.jpg)
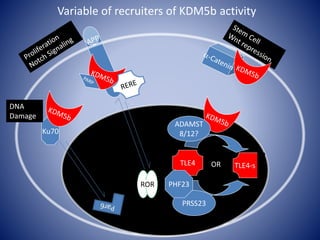
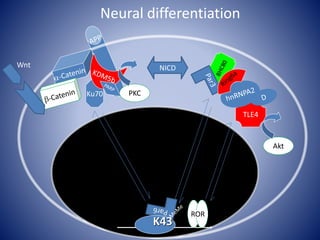
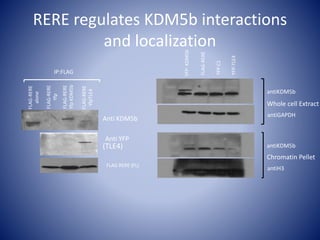
![Linking biochemistry to biological
properties-Future directions
1. Define relationship between KDM5 and
cell signaling (MS sequencing and
verification)
2. Define the cell biology that is altered by
modulation of this system (Post injury
NSCs and iPSCs [NSCs v skin])
3. What is the role of these proteins in
both injury and recovery (mouse
models)](https://image.slidesharecdn.com/epigeneticdrugs-140817111058-phpapp02/85/KDM5-epigenetic-modifiers-as-a-focus-for-drug-discovery-30-320.jpg)
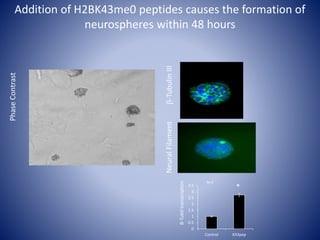
![Linking biochemistry to biological
properties-Future directions
1. Define relationship between KDM5 and
cell signaling (MS sequencing and
verification)
2. Define the cell biology that is altered by
modulation of this system (Post injury
NSCs and iPSCs [NSCs v skin])
3. What is the role of these proteins in
both injury and recovery (mouse
models)](https://image.slidesharecdn.com/epigeneticdrugs-140817111058-phpapp02/85/KDM5-epigenetic-modifiers-as-a-focus-for-drug-discovery-41-320.jpg)