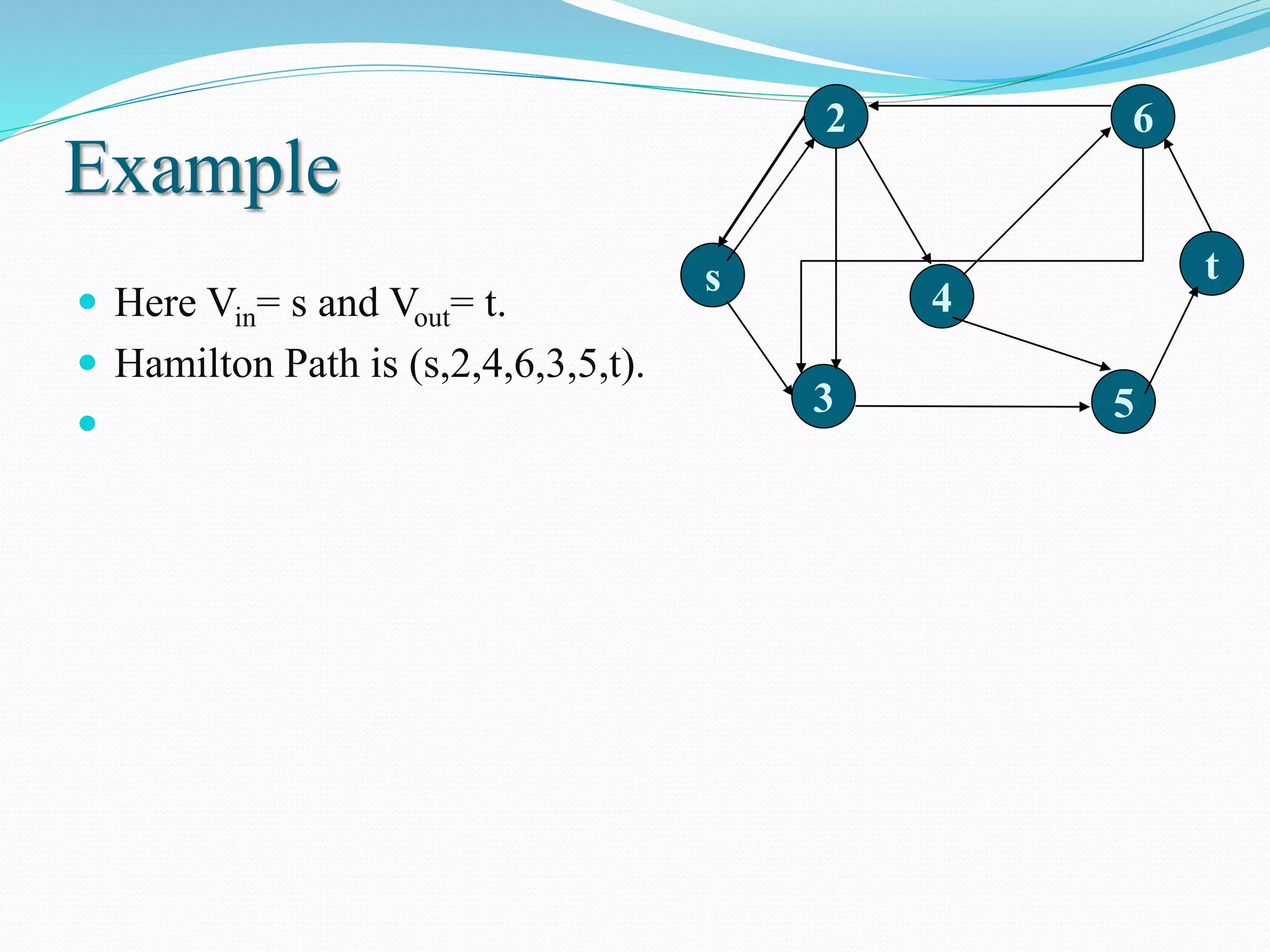
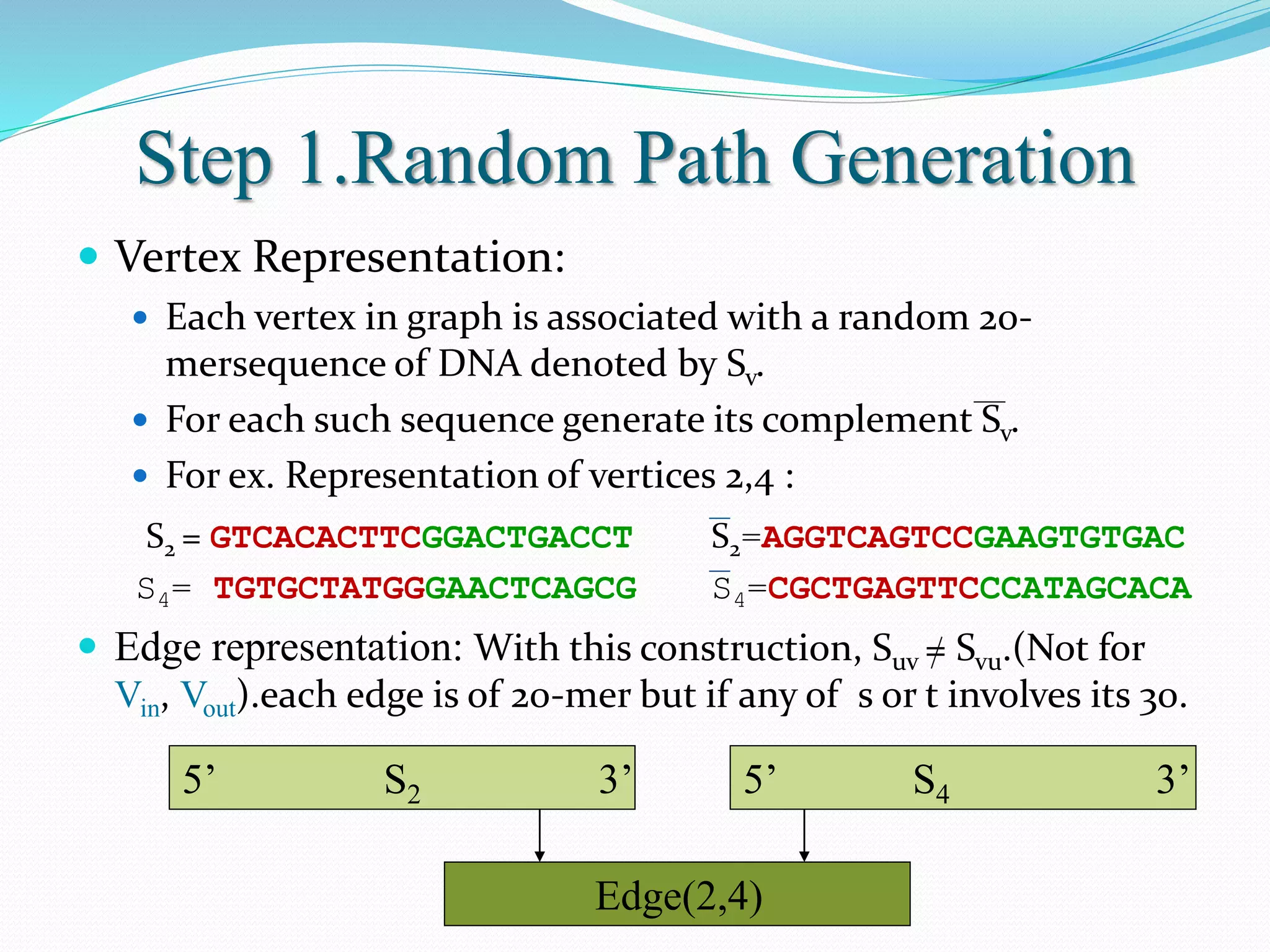
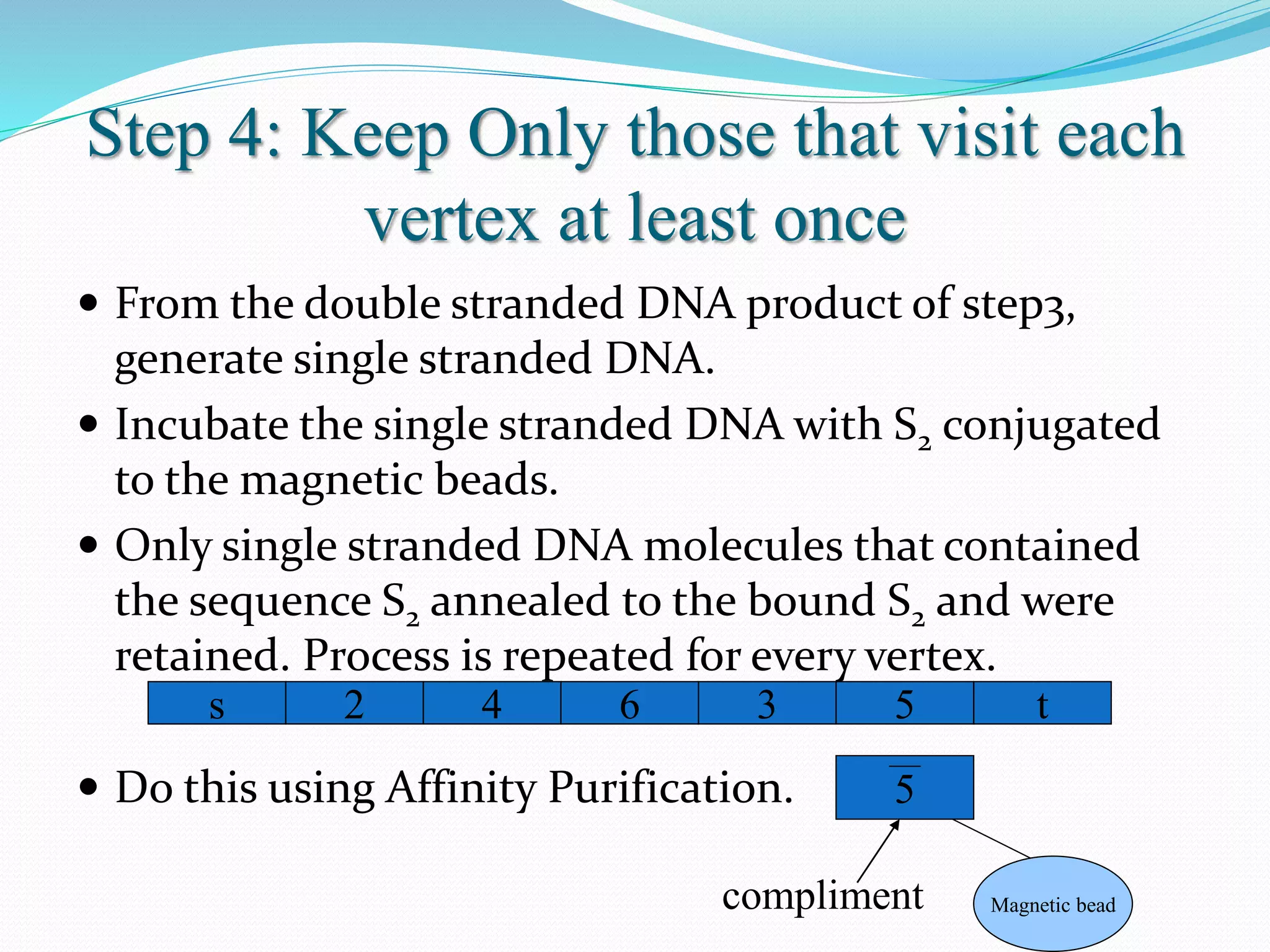
This document discusses DNA computing, including its potential advantages over traditional silicon-based computing. It summarizes Adleman's 1994 experiment solving the Hamiltonian path problem using DNA operations like annealing, separation by length/sequence, and affinity purification. While DNA computing offers massive parallelism and low power usage, challenges include error rates increasing with problem size and the difficulty of transmitting information between DNA strands. The future of DNA computing may include applications in encryption, genetic programming, and logistics optimization, though general-purpose DNA computers are unlikely.