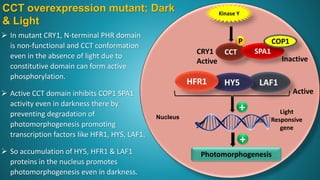
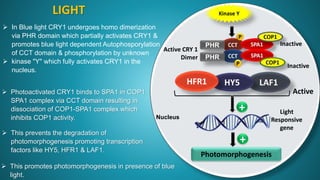
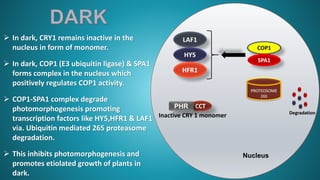
1. Cryptochrome is a major blue light photoreceptor in plants that regulates processes like photomorphogenesis, circadian rhythms, and flowering.
2. There are three types of cryptochrome found in Arabidopsis - CRY1, CRY2, and CRY3. CRY1 and CRY2 are involved in photomorphogenesis while CRY3 functions in DNA repair in mitochondria and chloroplasts.
3. Cryptochrome has two domains - an amino-terminal photolyase homology region that binds chromophores like FAD and perceives blue light, and a carboxy-terminal output domain that transduces the light signal.