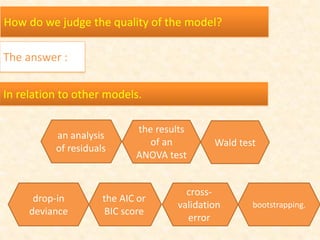
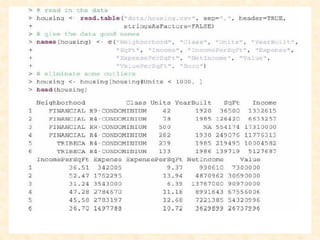
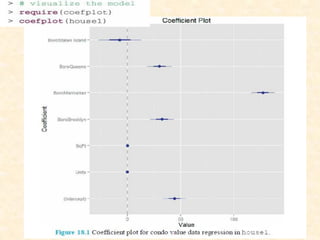
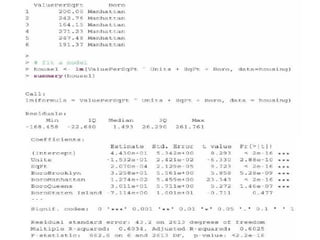
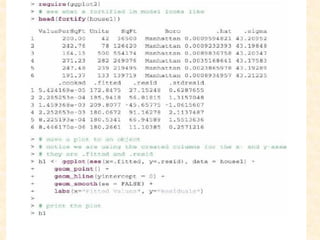
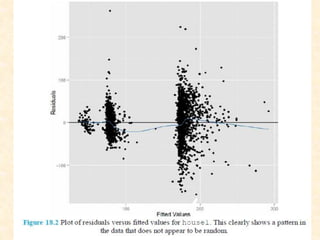
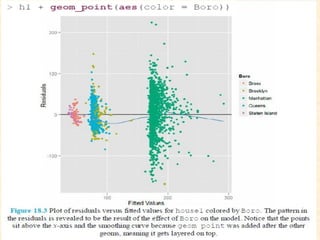
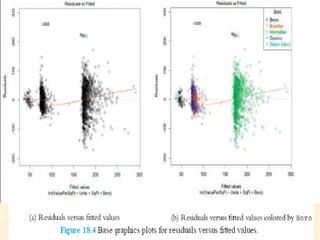
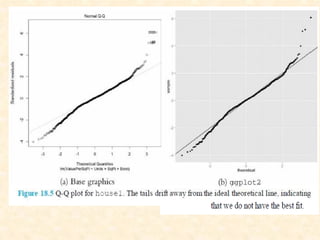
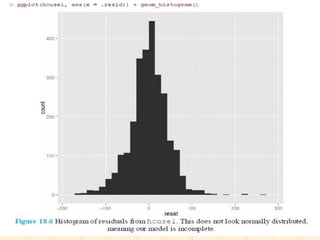
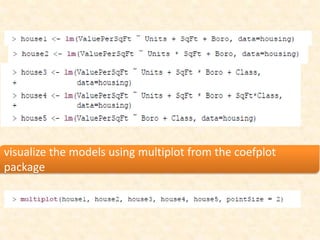
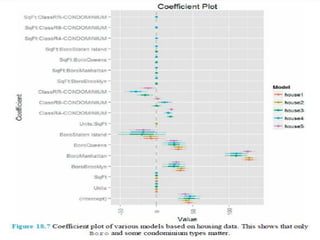
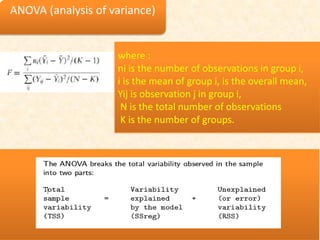
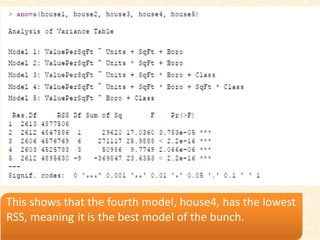
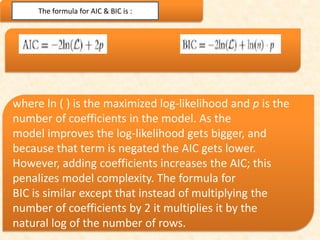
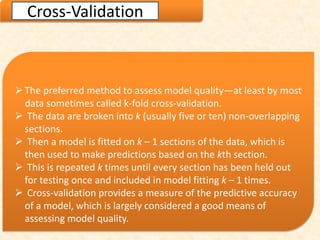
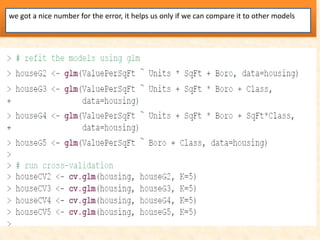
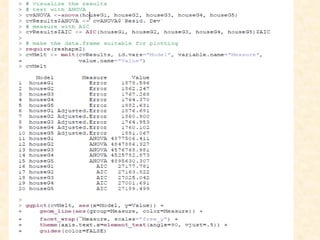
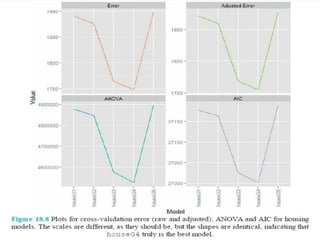
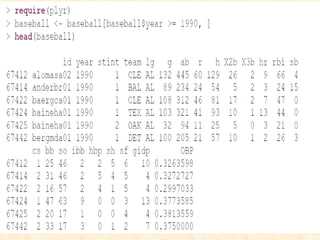
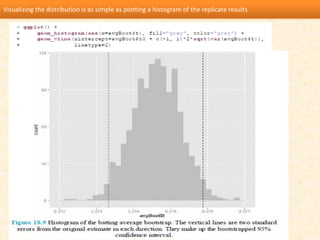
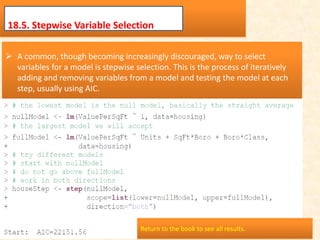
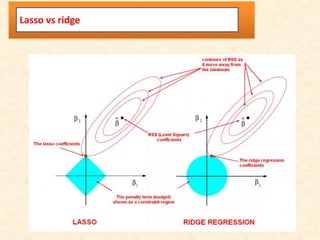
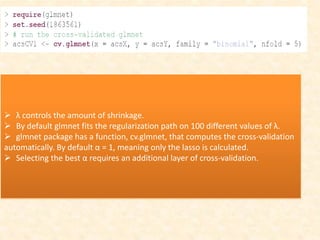
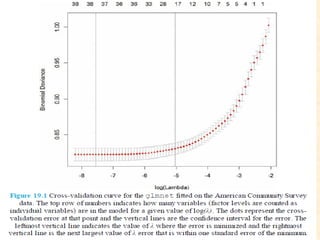
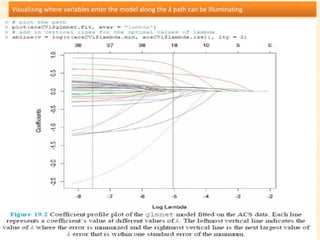
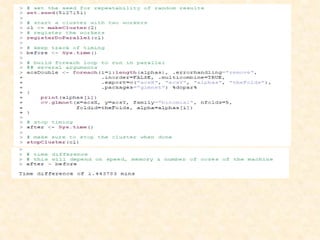
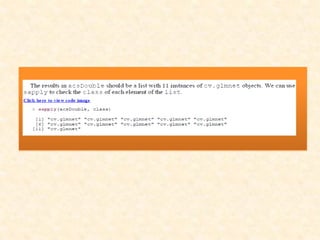
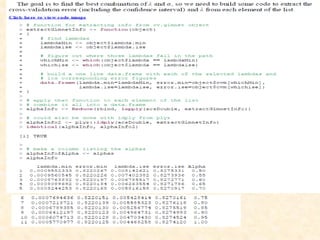
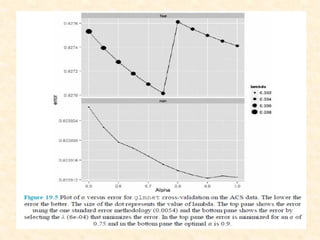
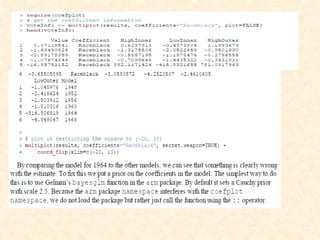
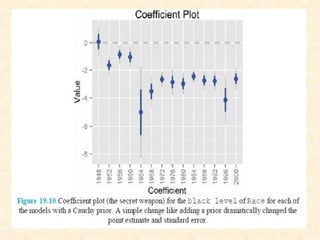
This document discusses techniques for evaluating and improving statistical models, including regularized regression methods. It covers residuals, Q-Q plots, histograms to evaluate model fit. It also discusses comparing models using ANOVA, AIC, BIC, cross-validation, bootstrapping. Regularization methods like lasso, ridge and elastic net are introduced. Parallel computing is used to more efficiently select hyperparameters for elastic net models.