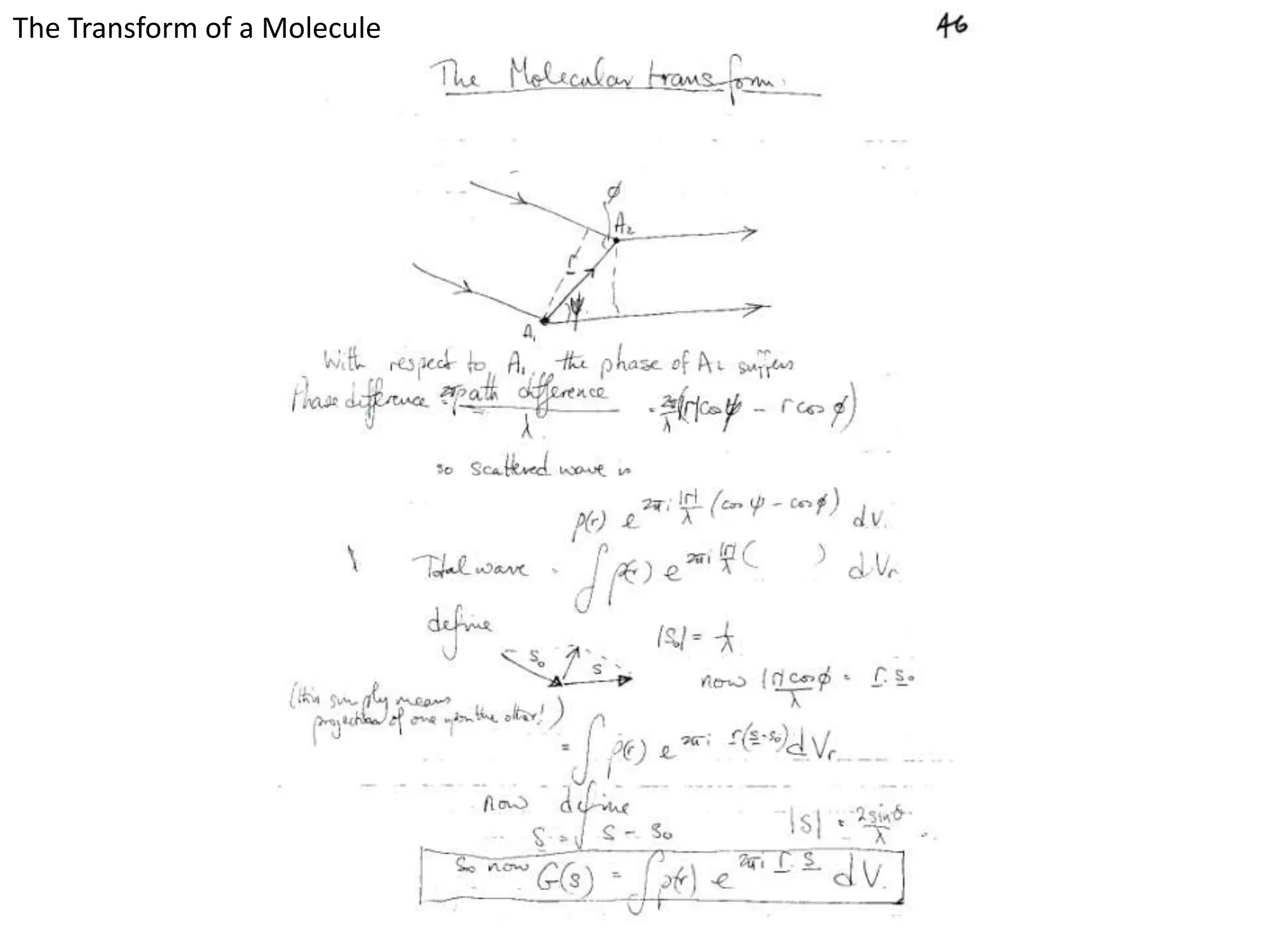
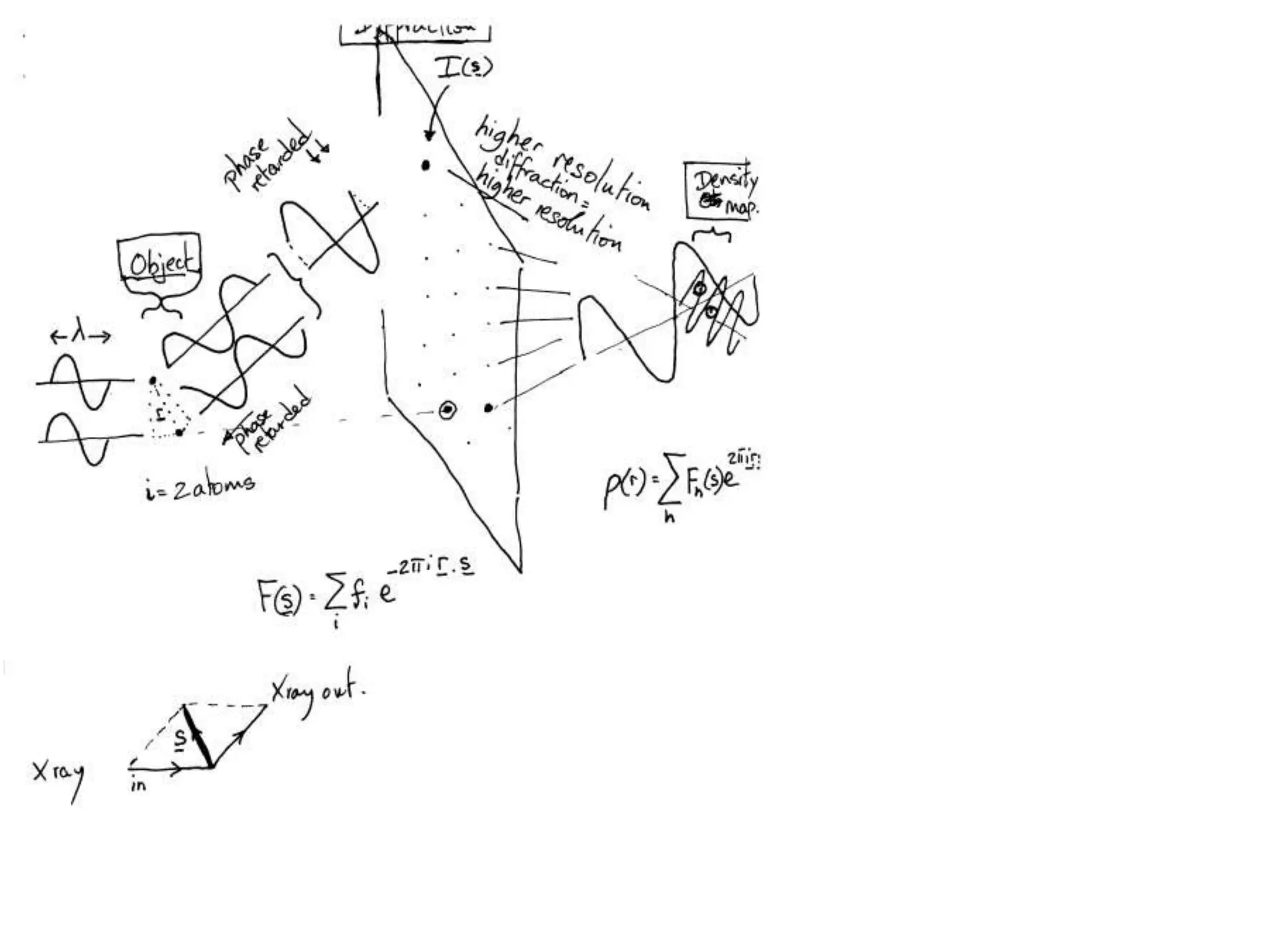
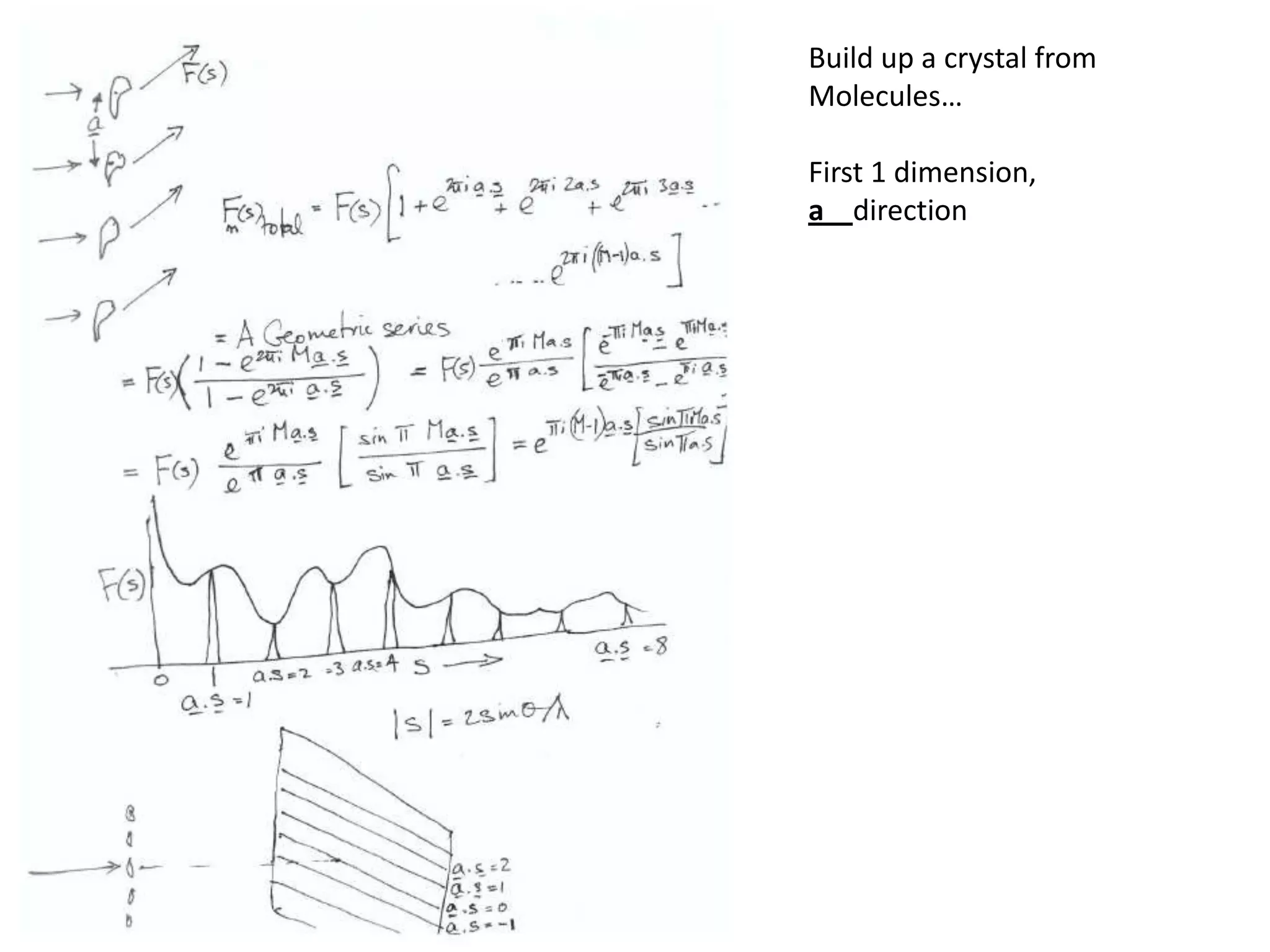
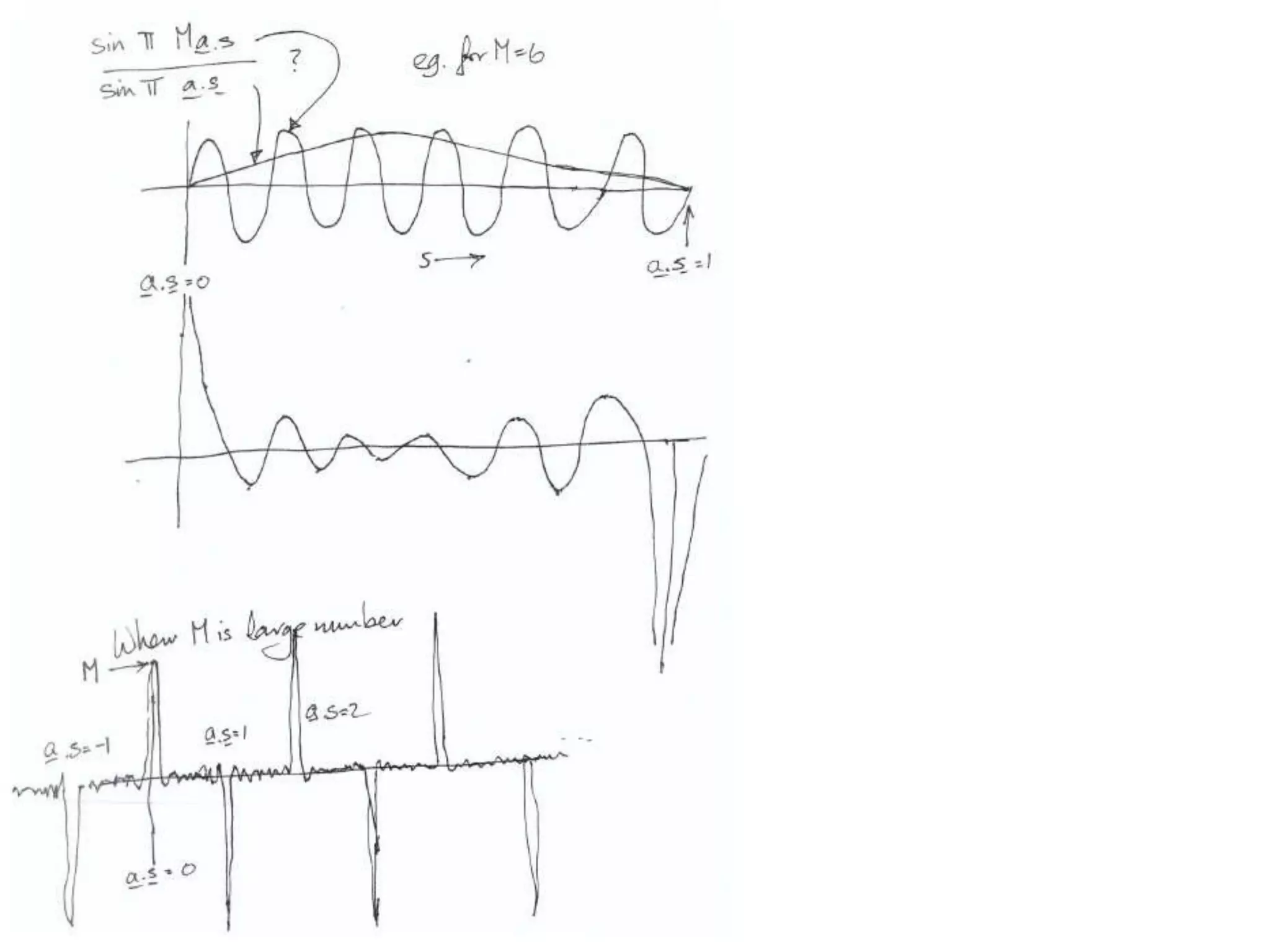
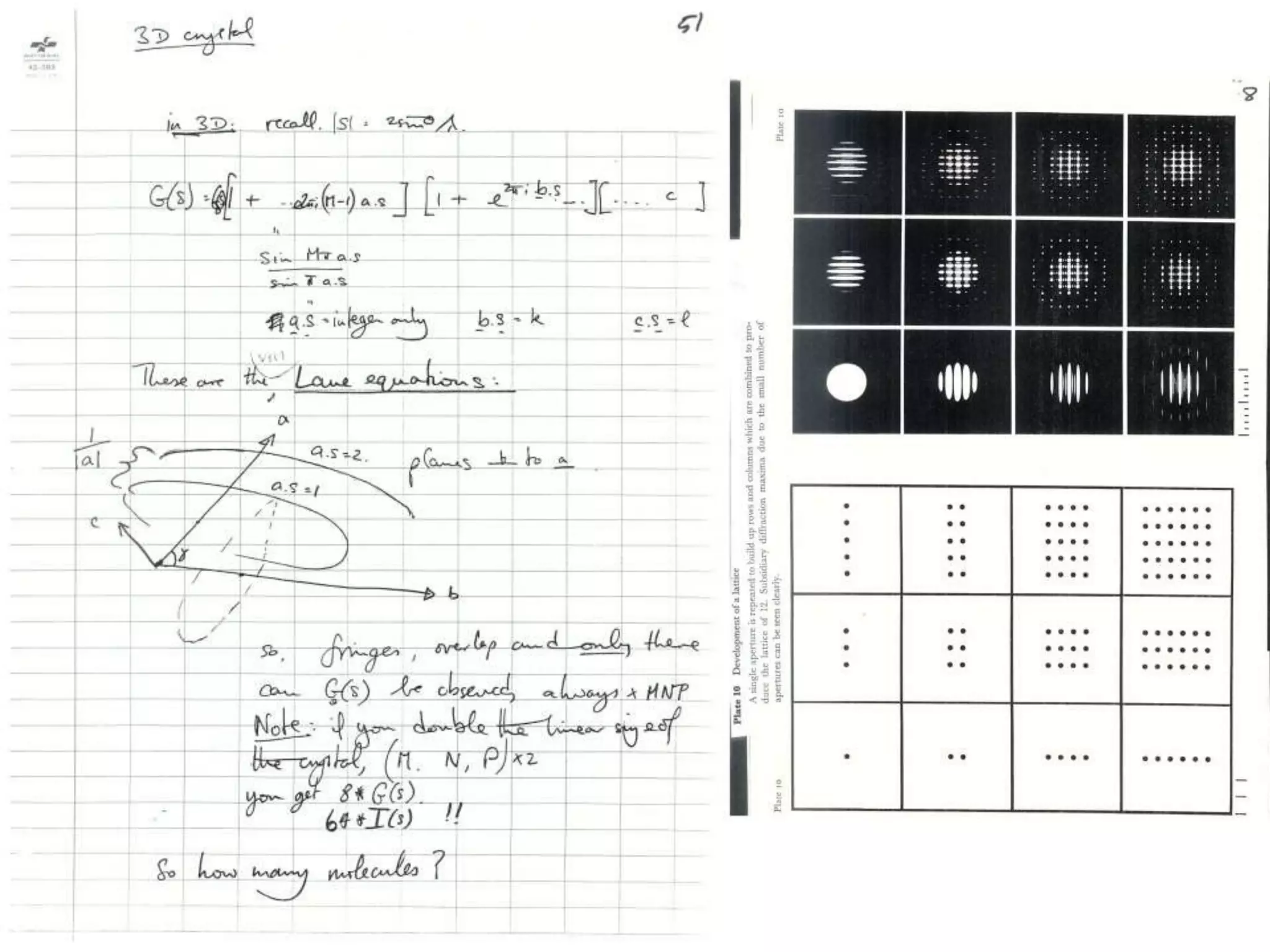
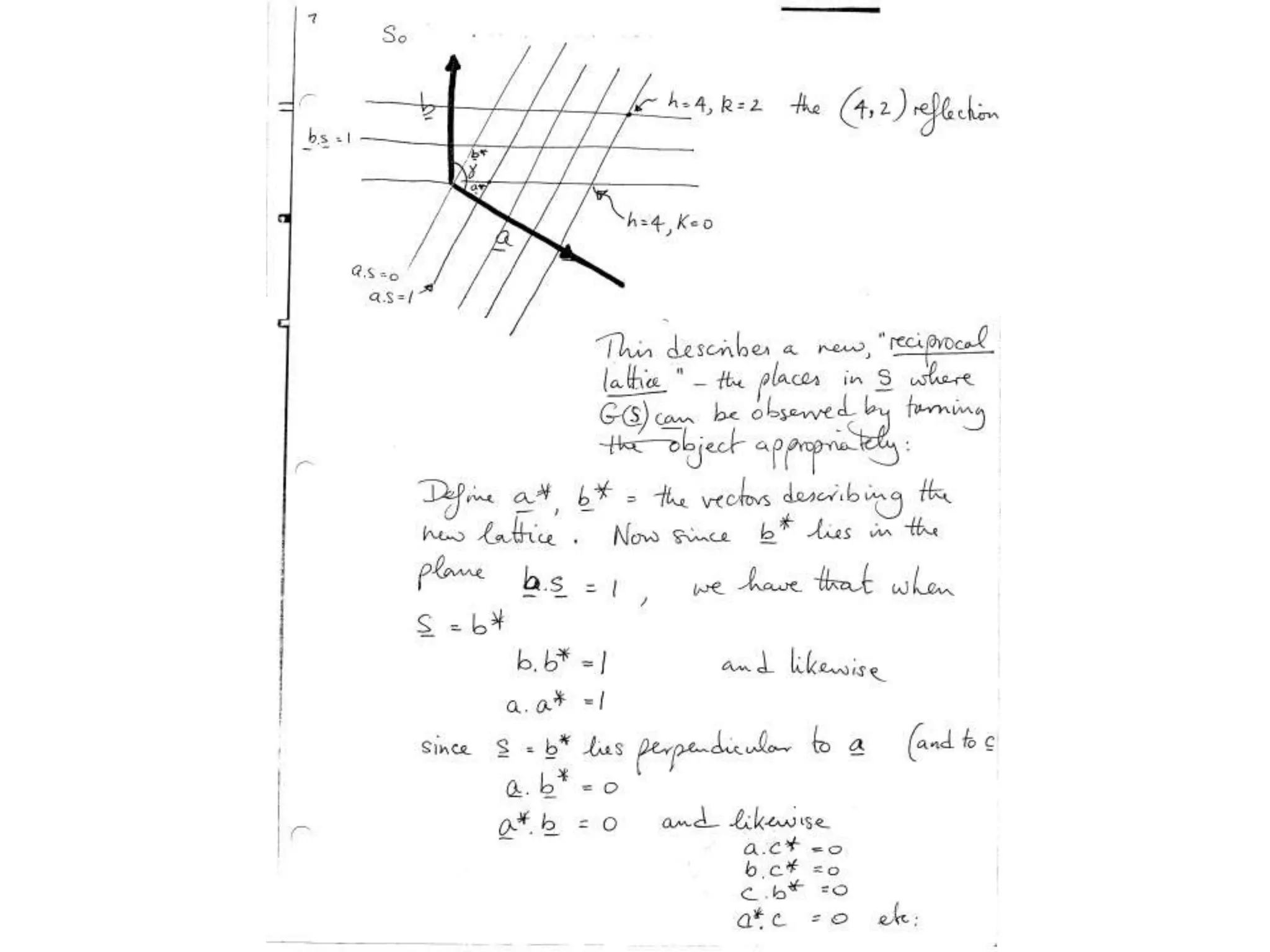
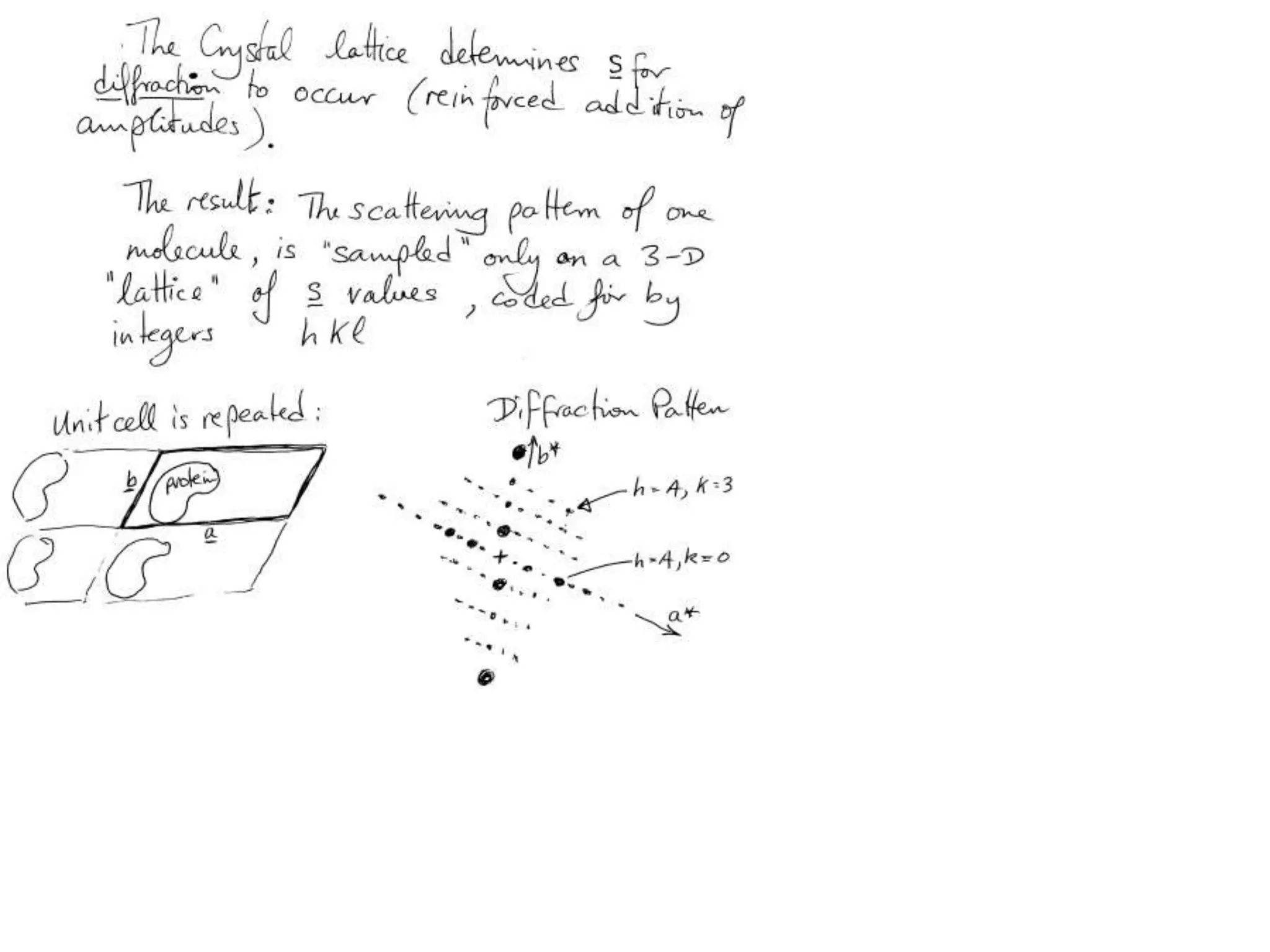
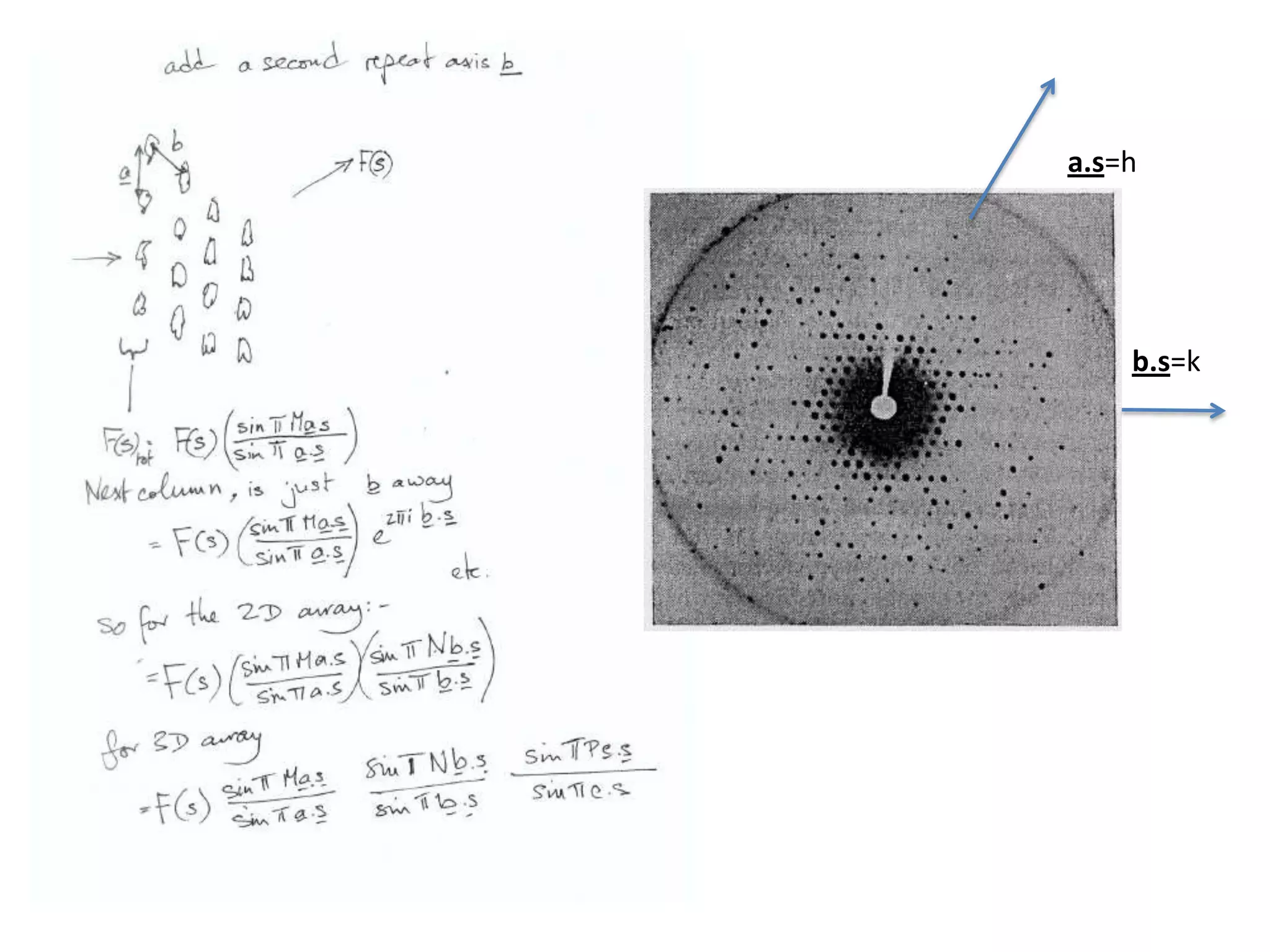
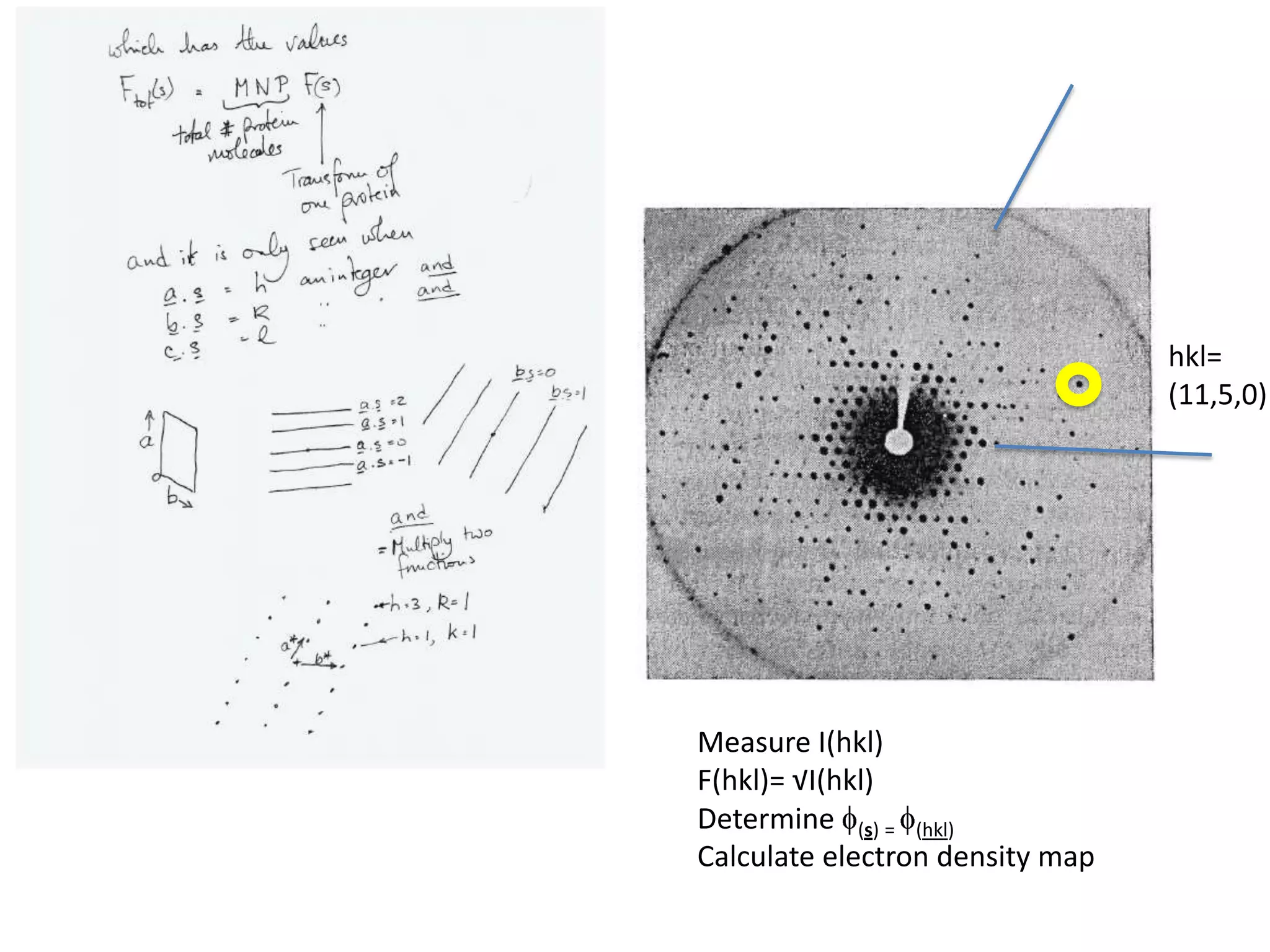
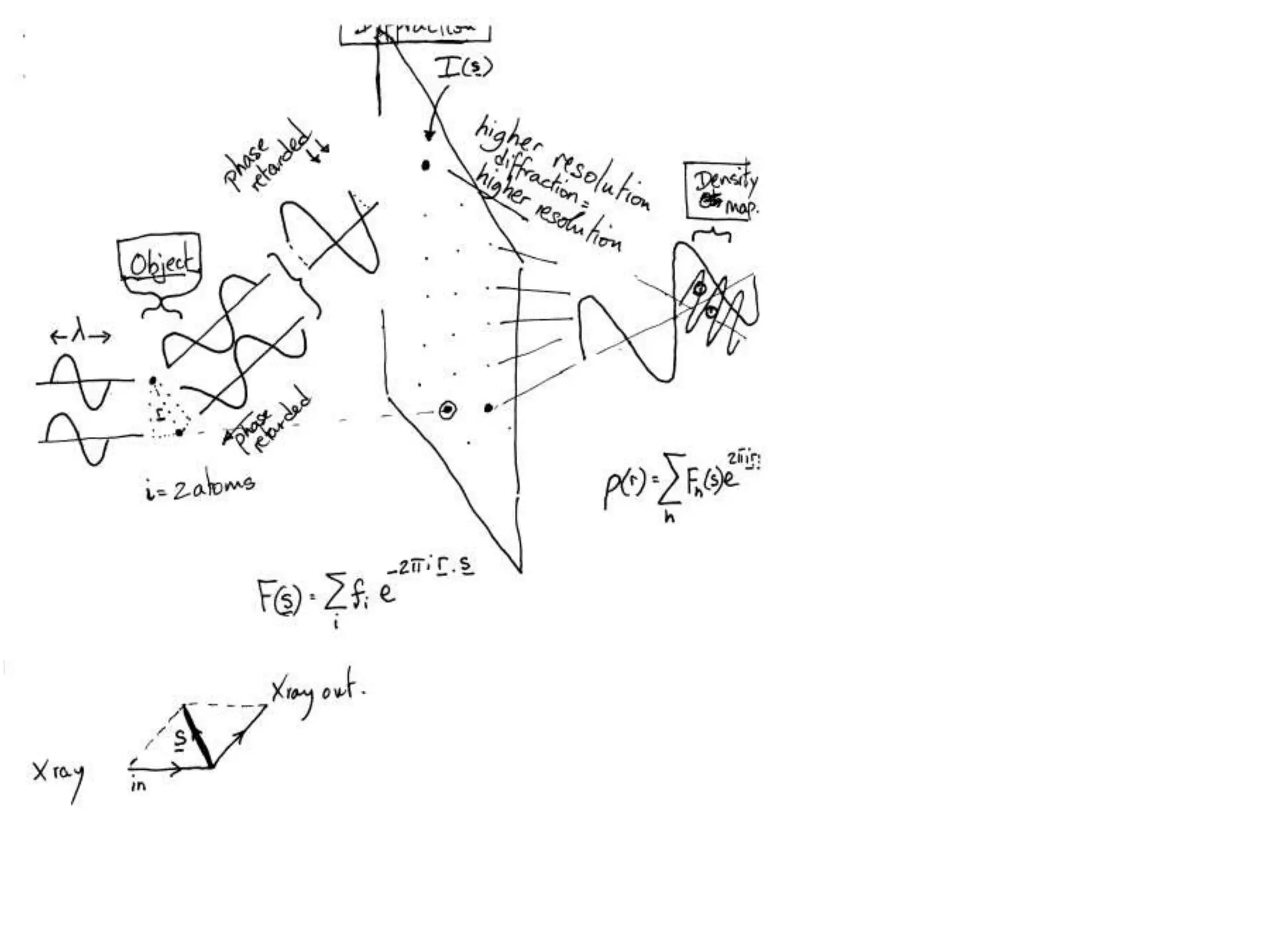
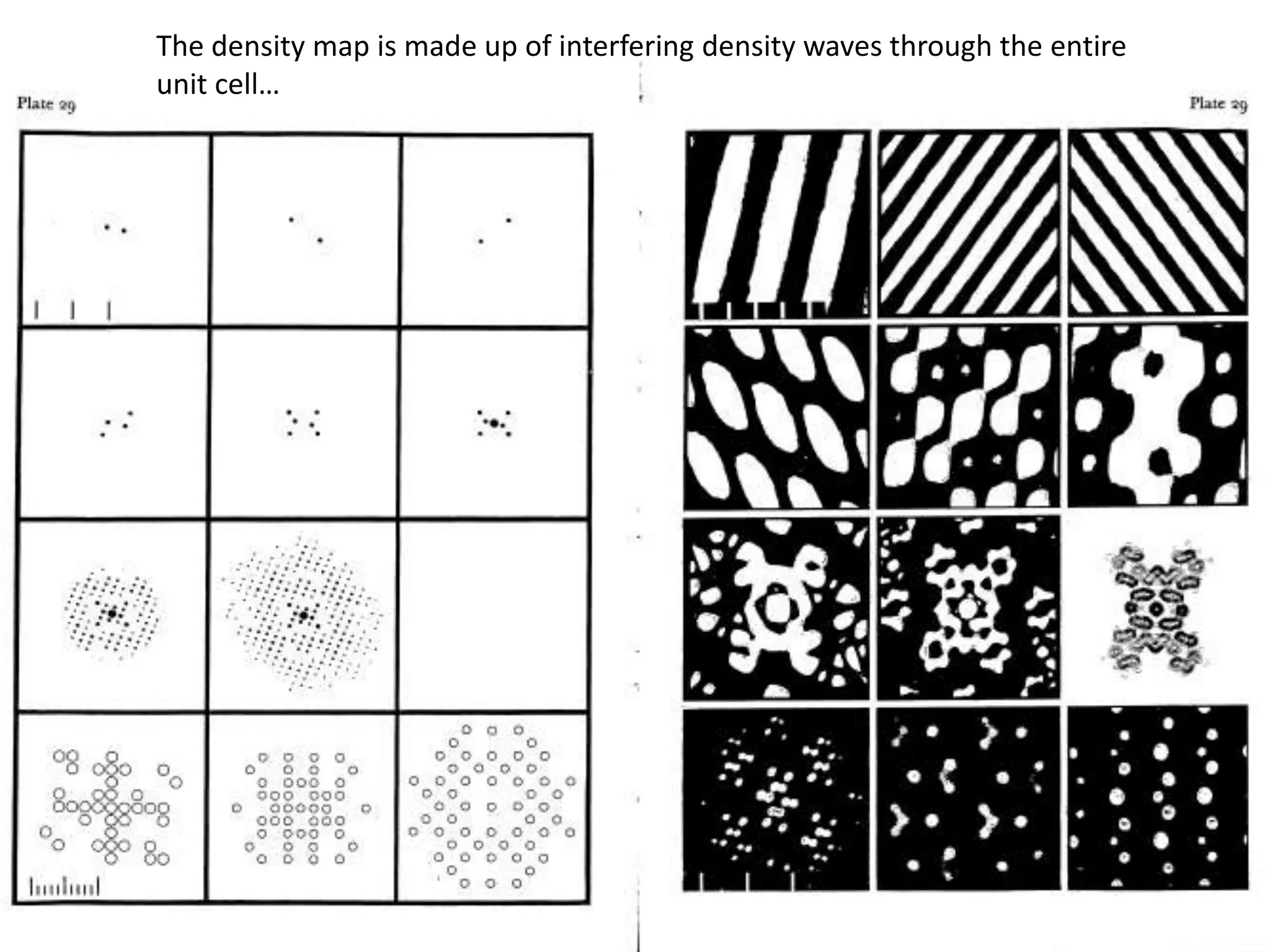
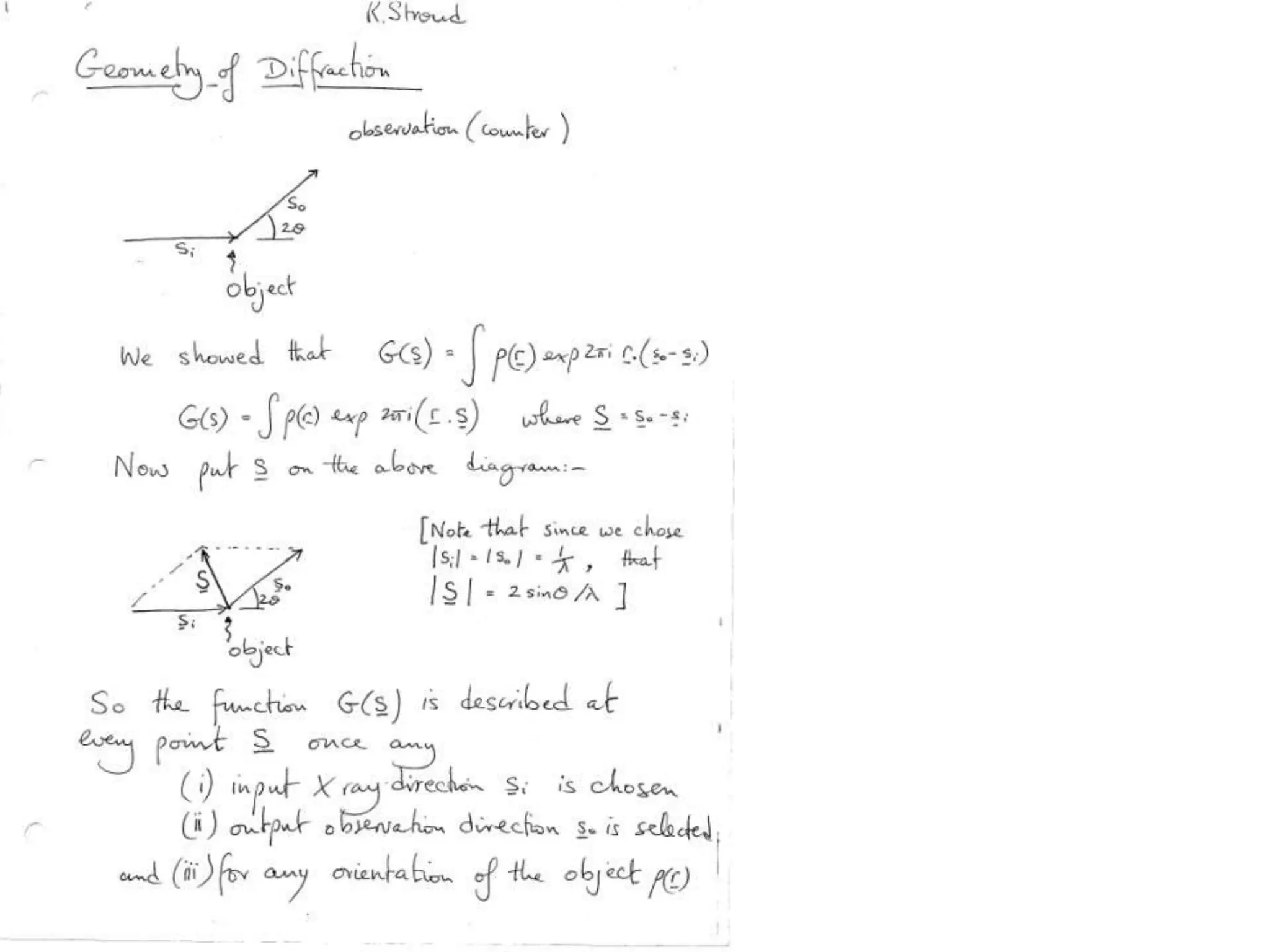
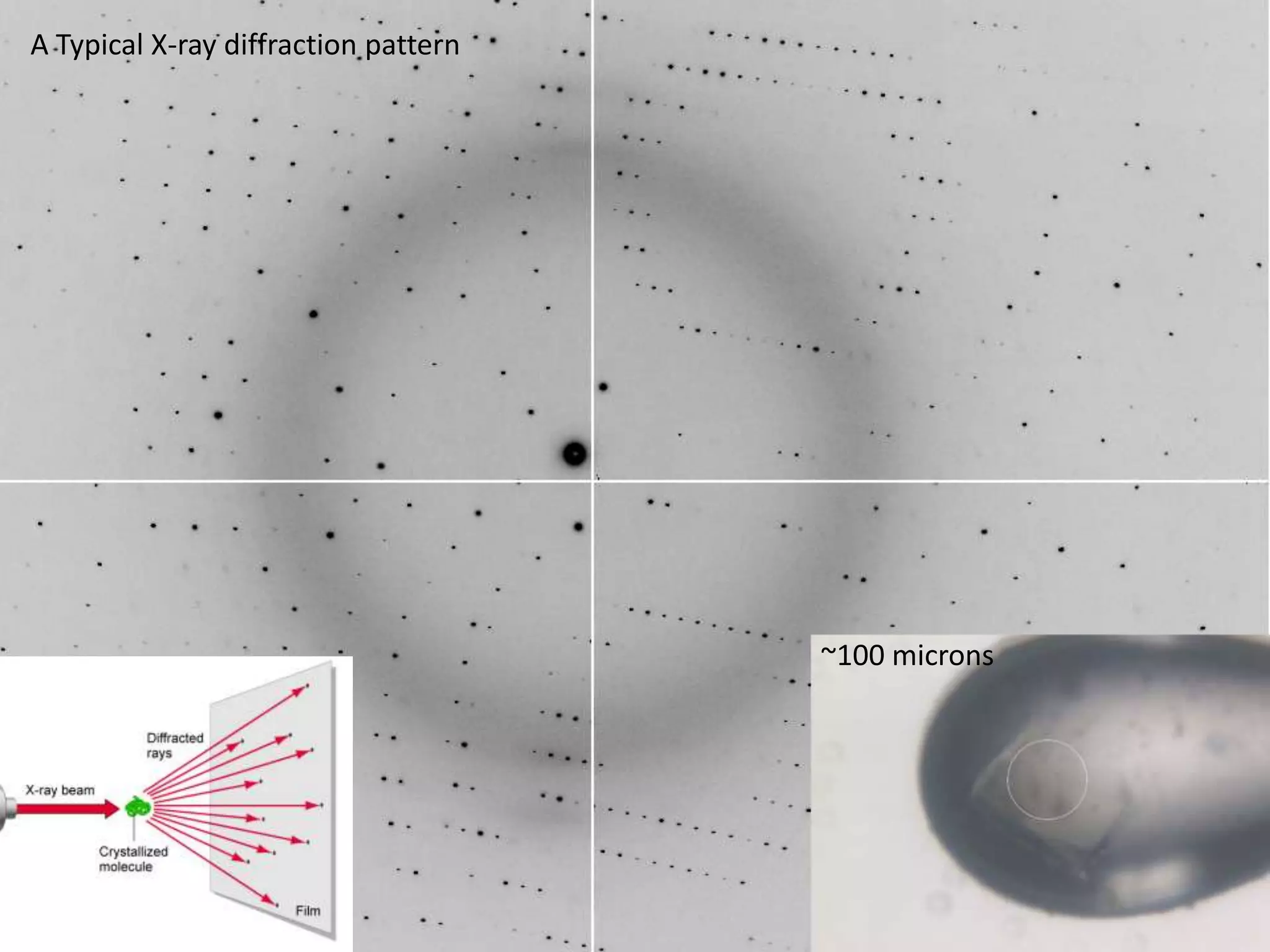
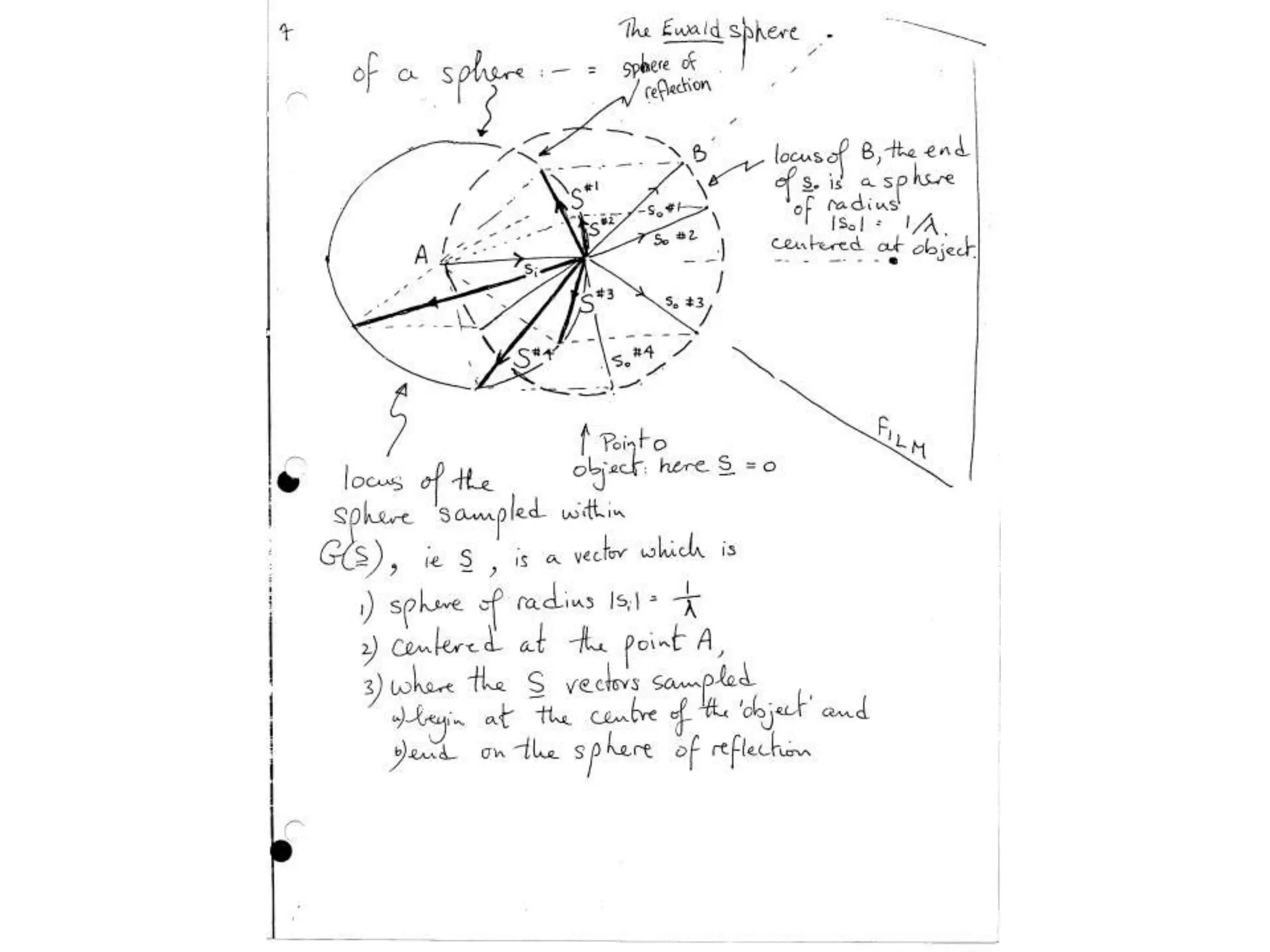
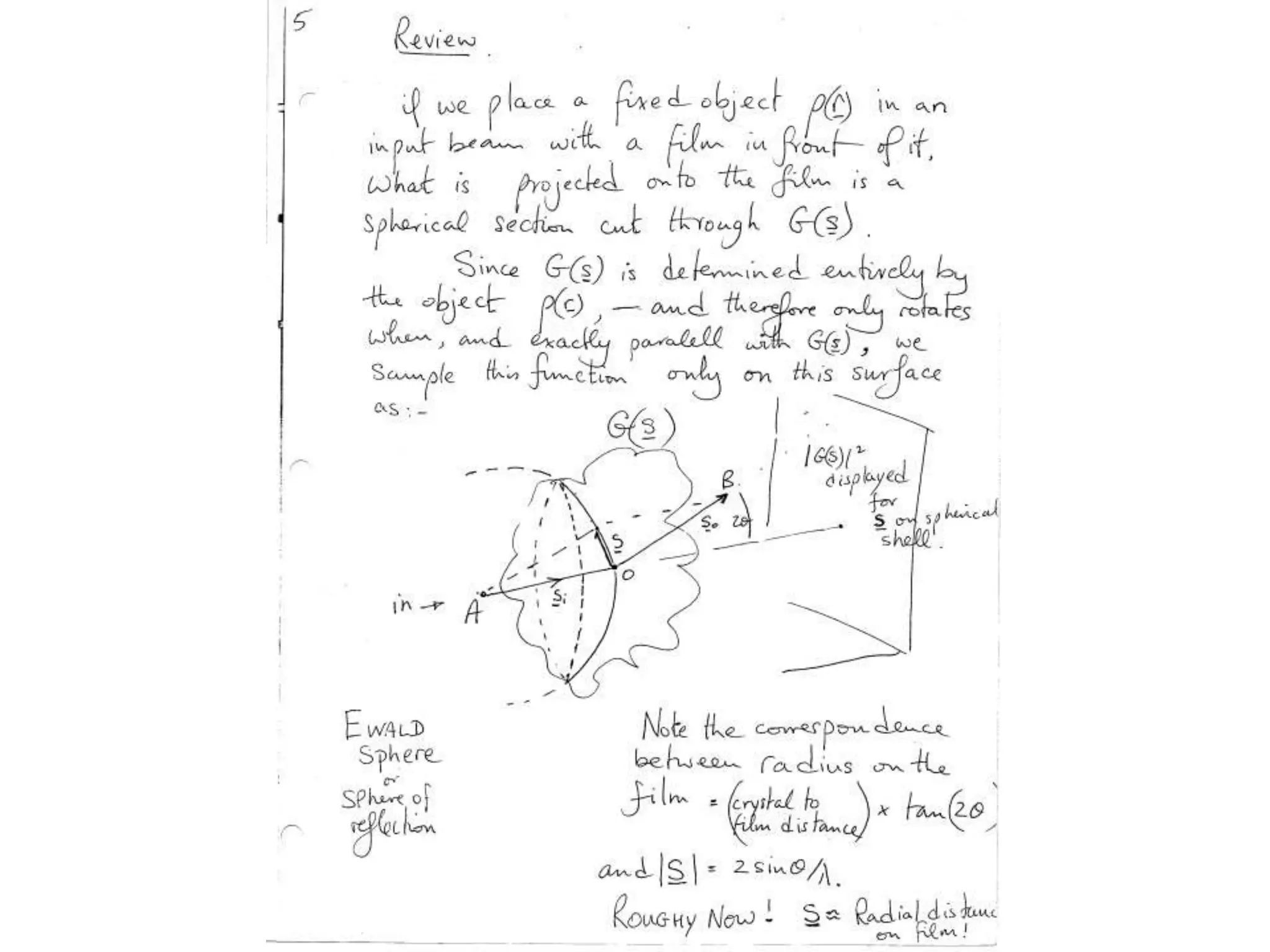
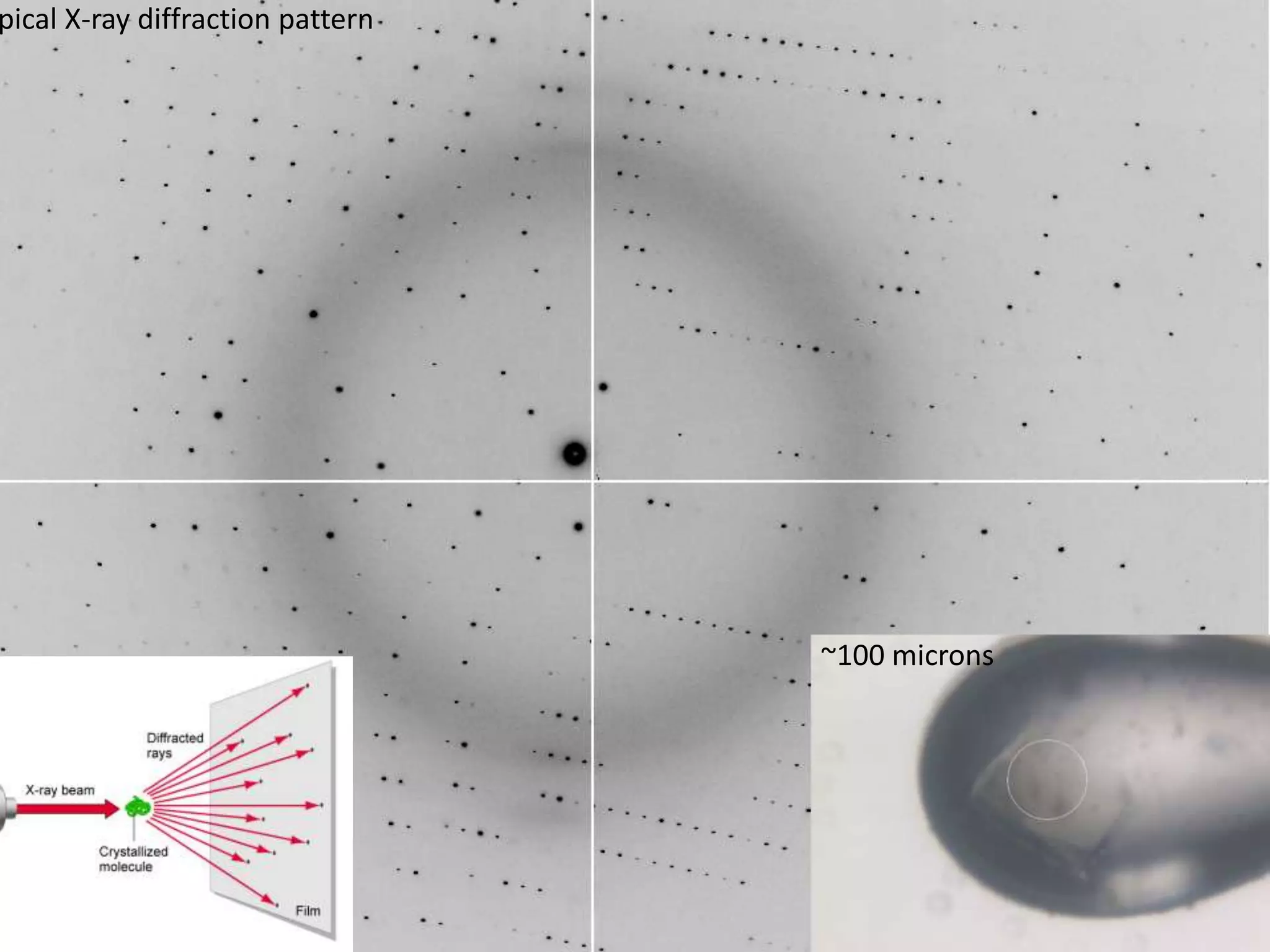
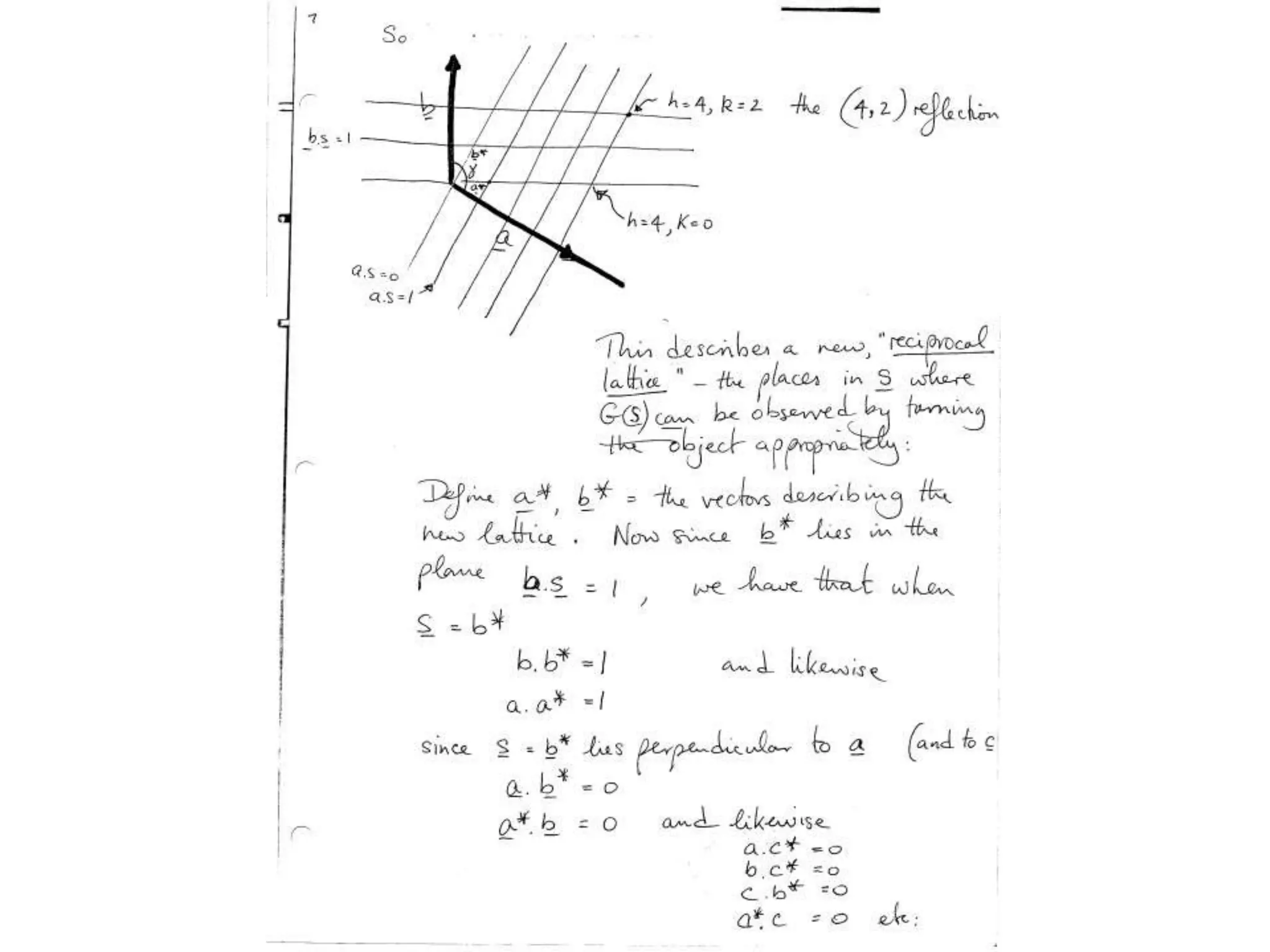
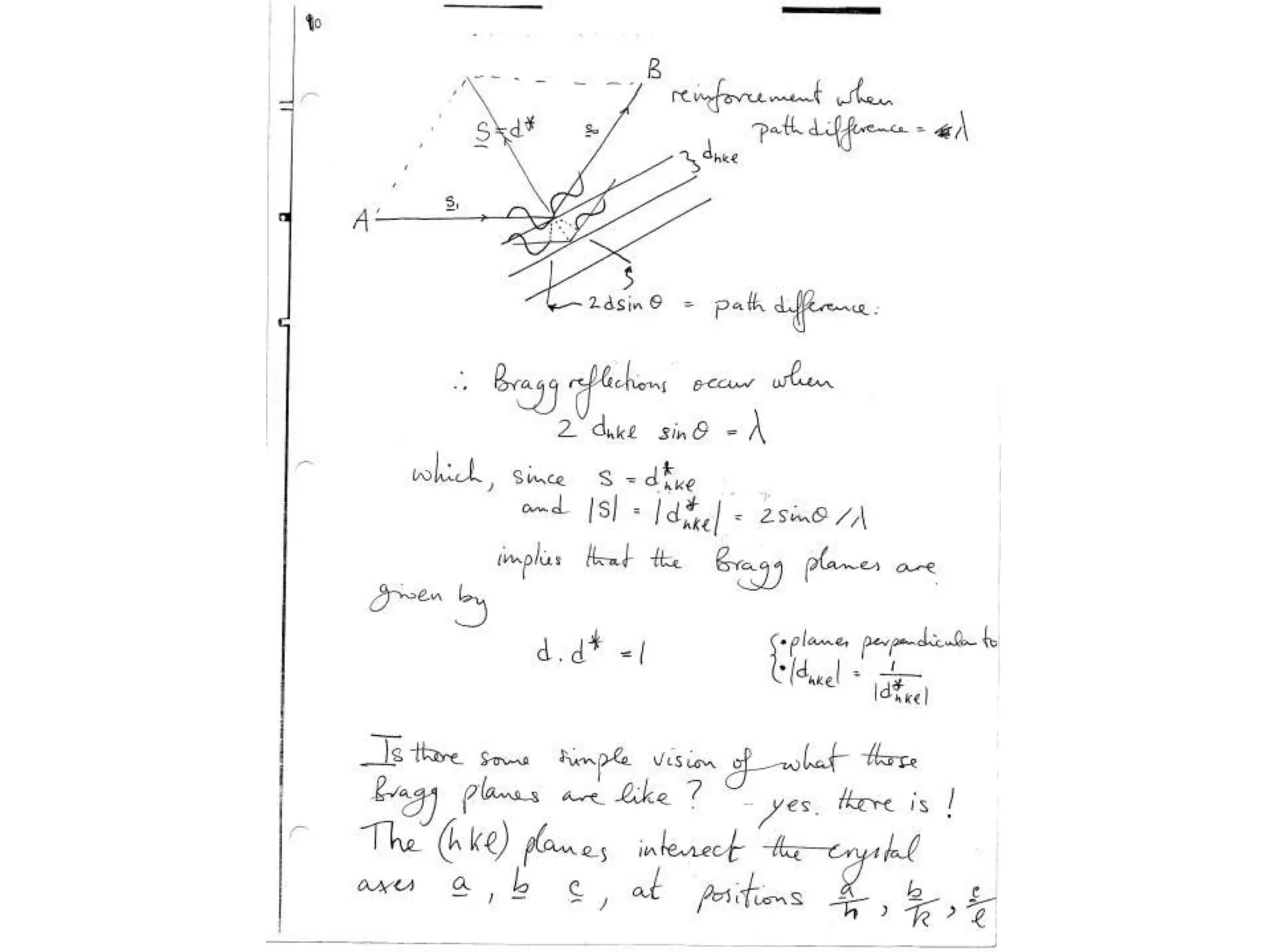
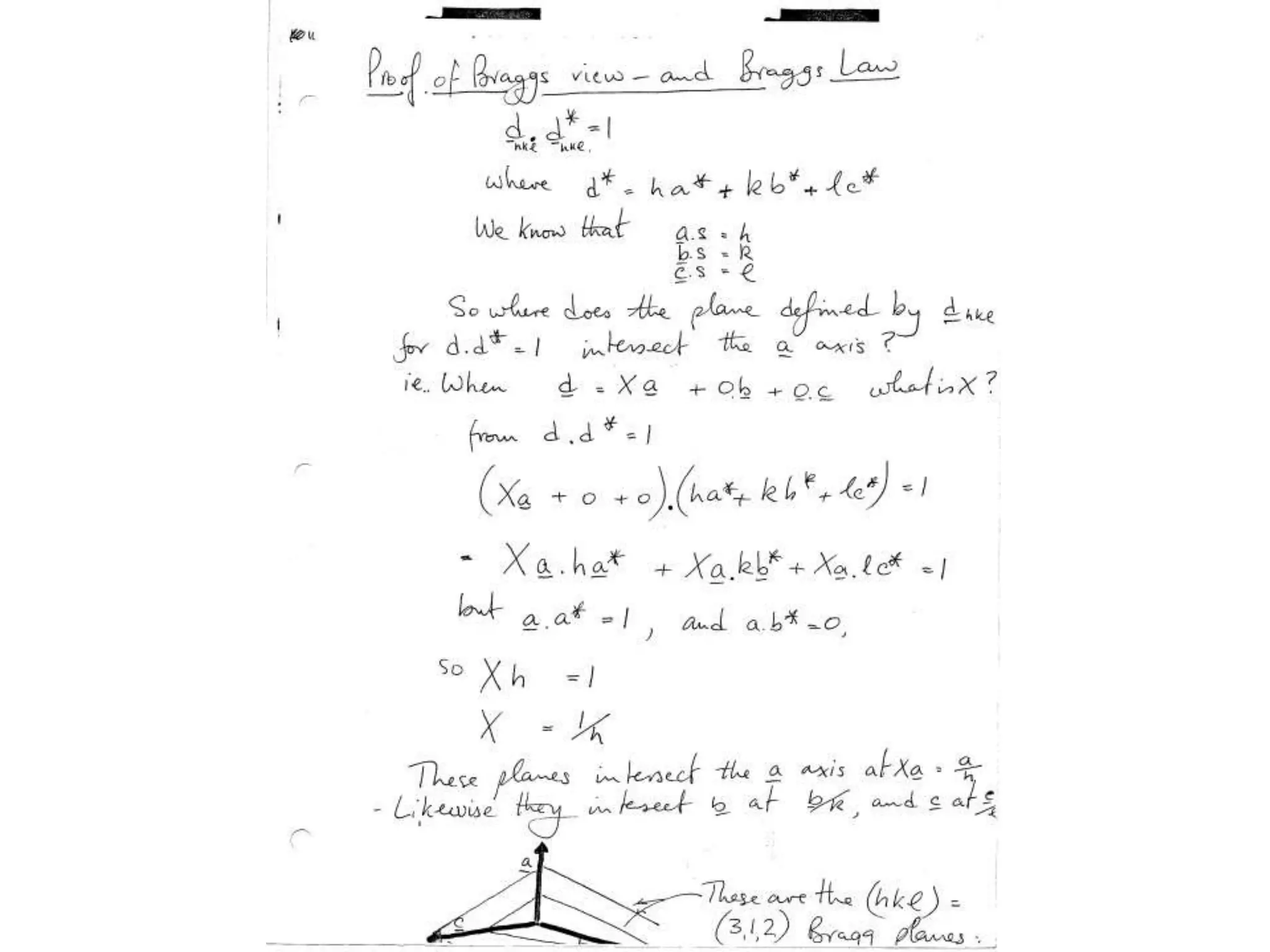
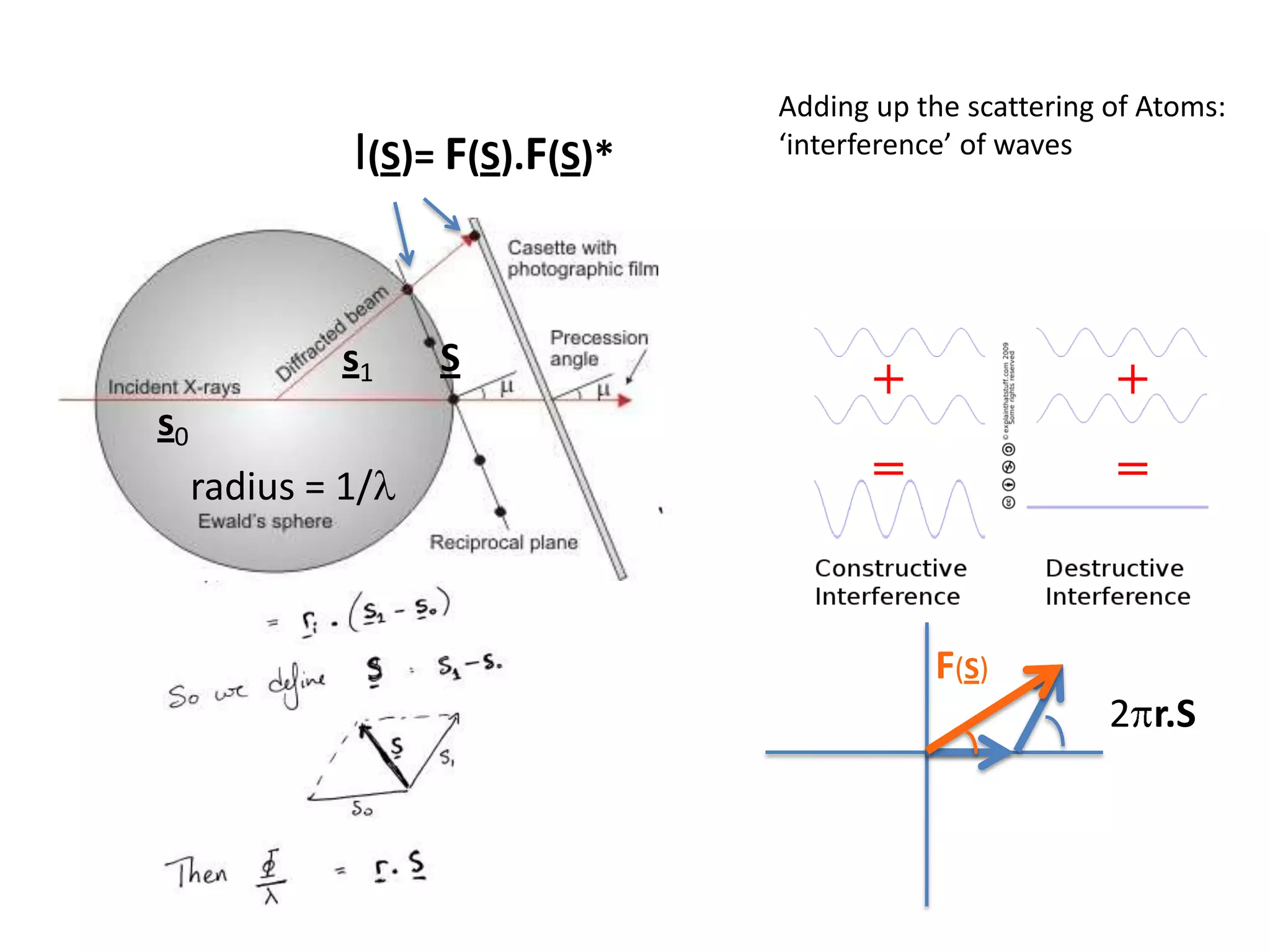
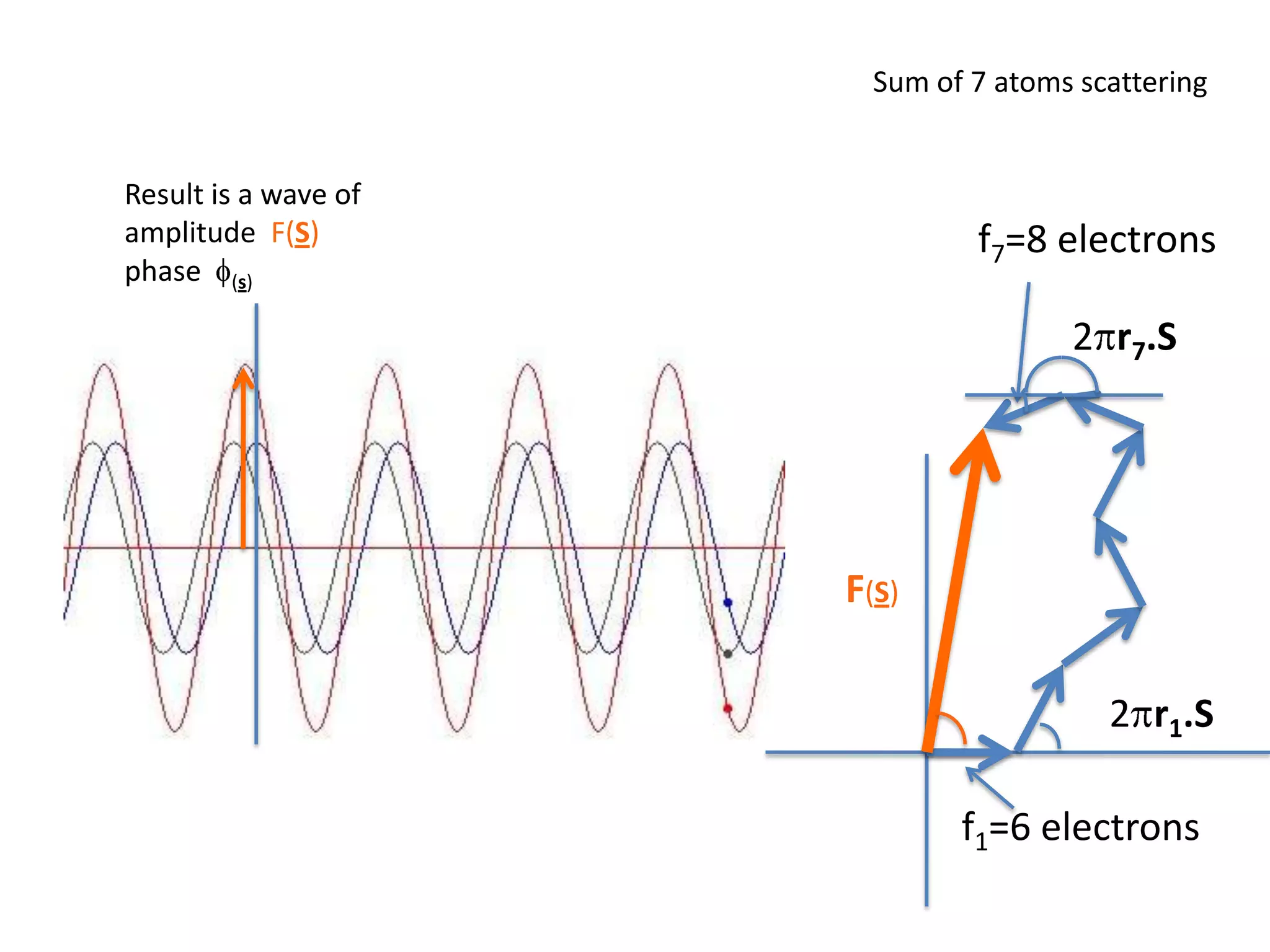
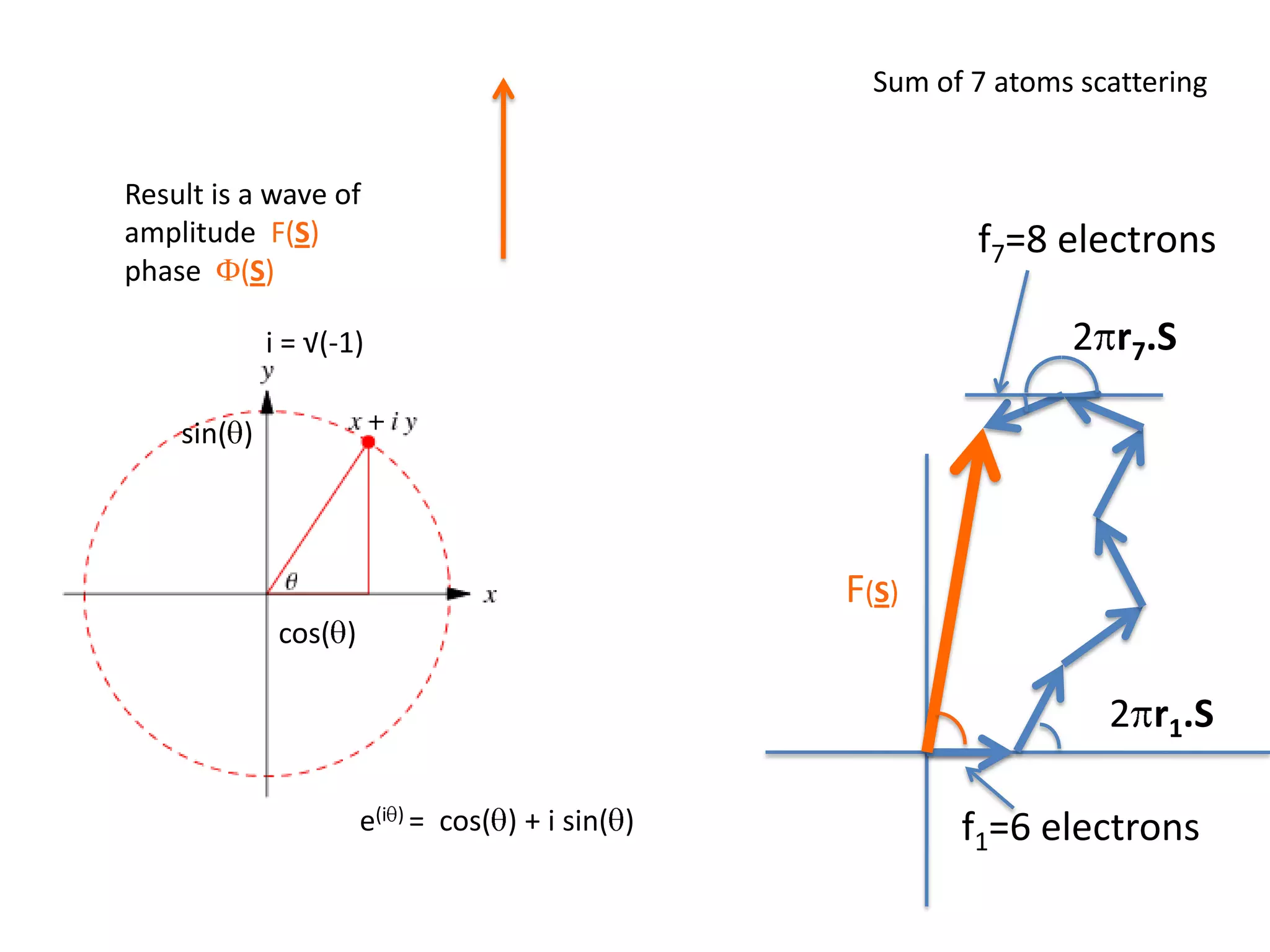
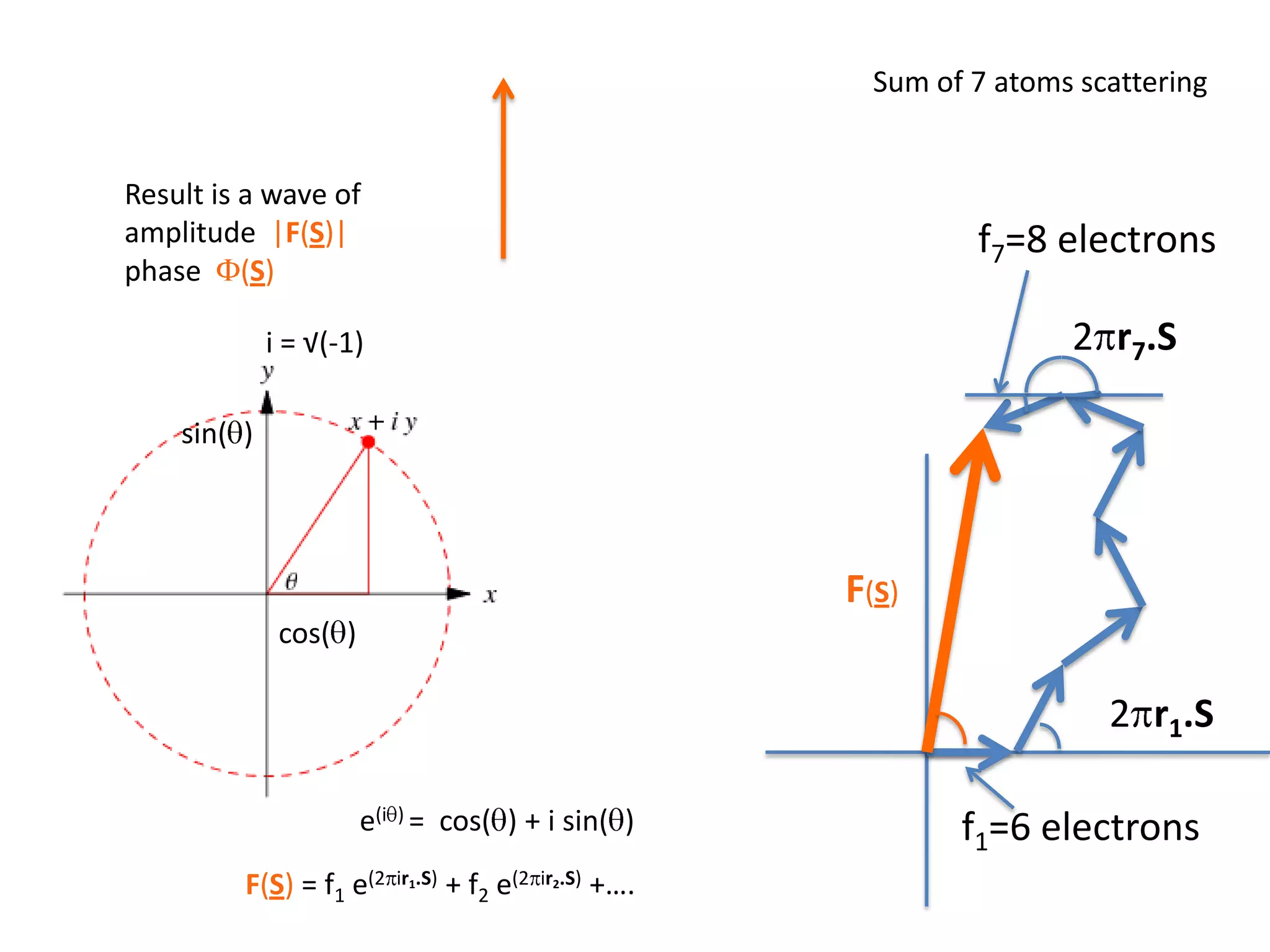
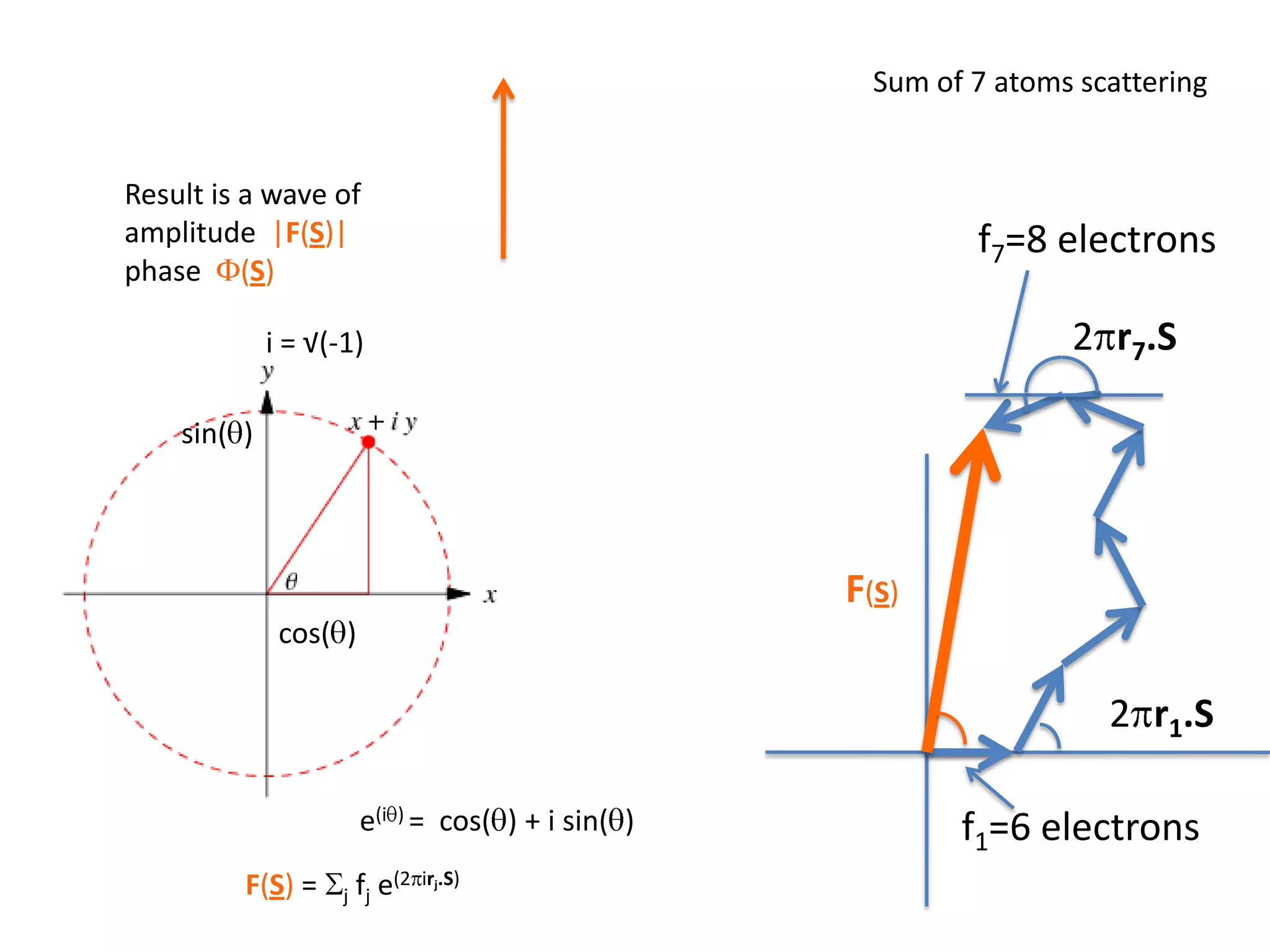
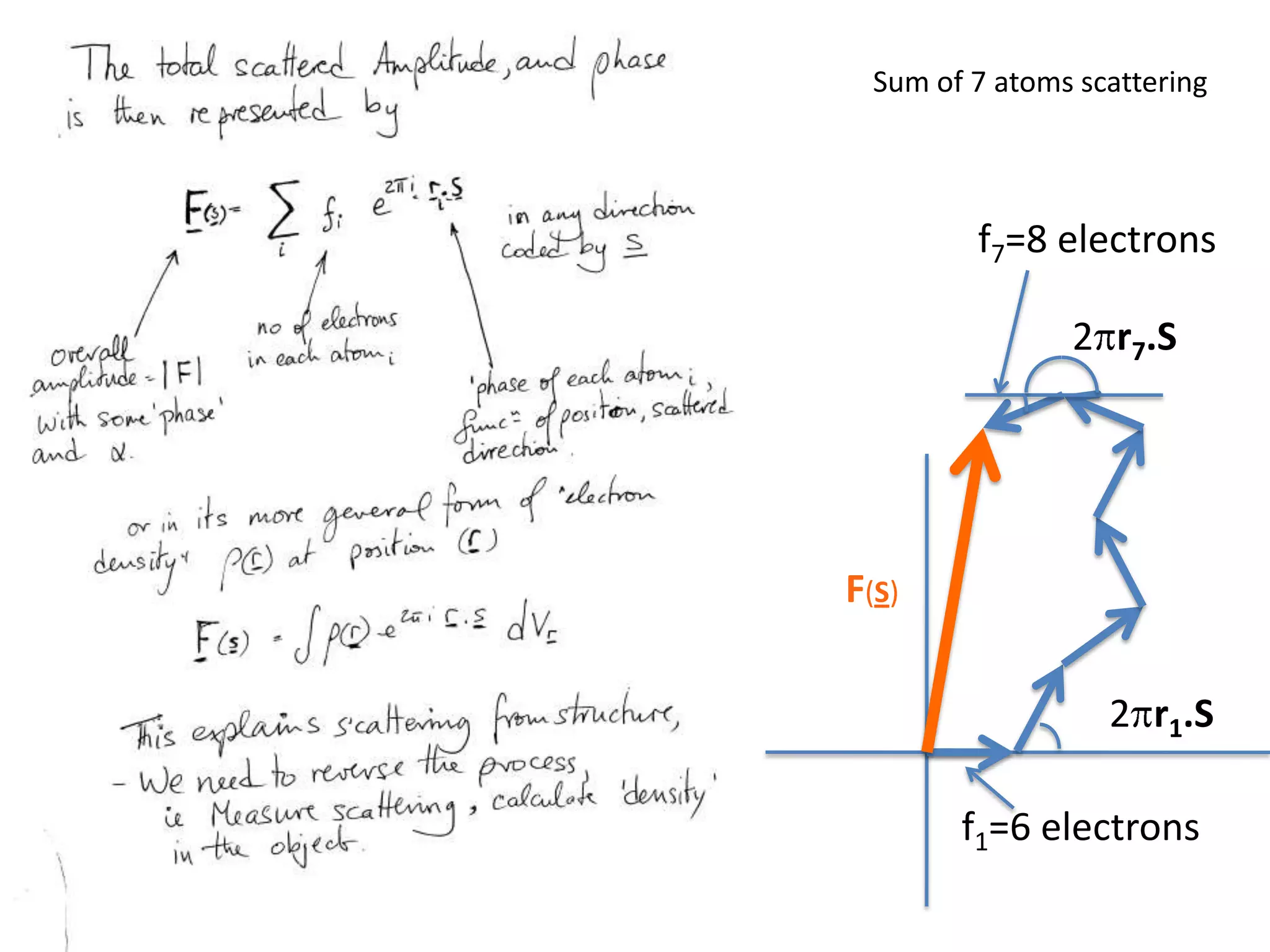
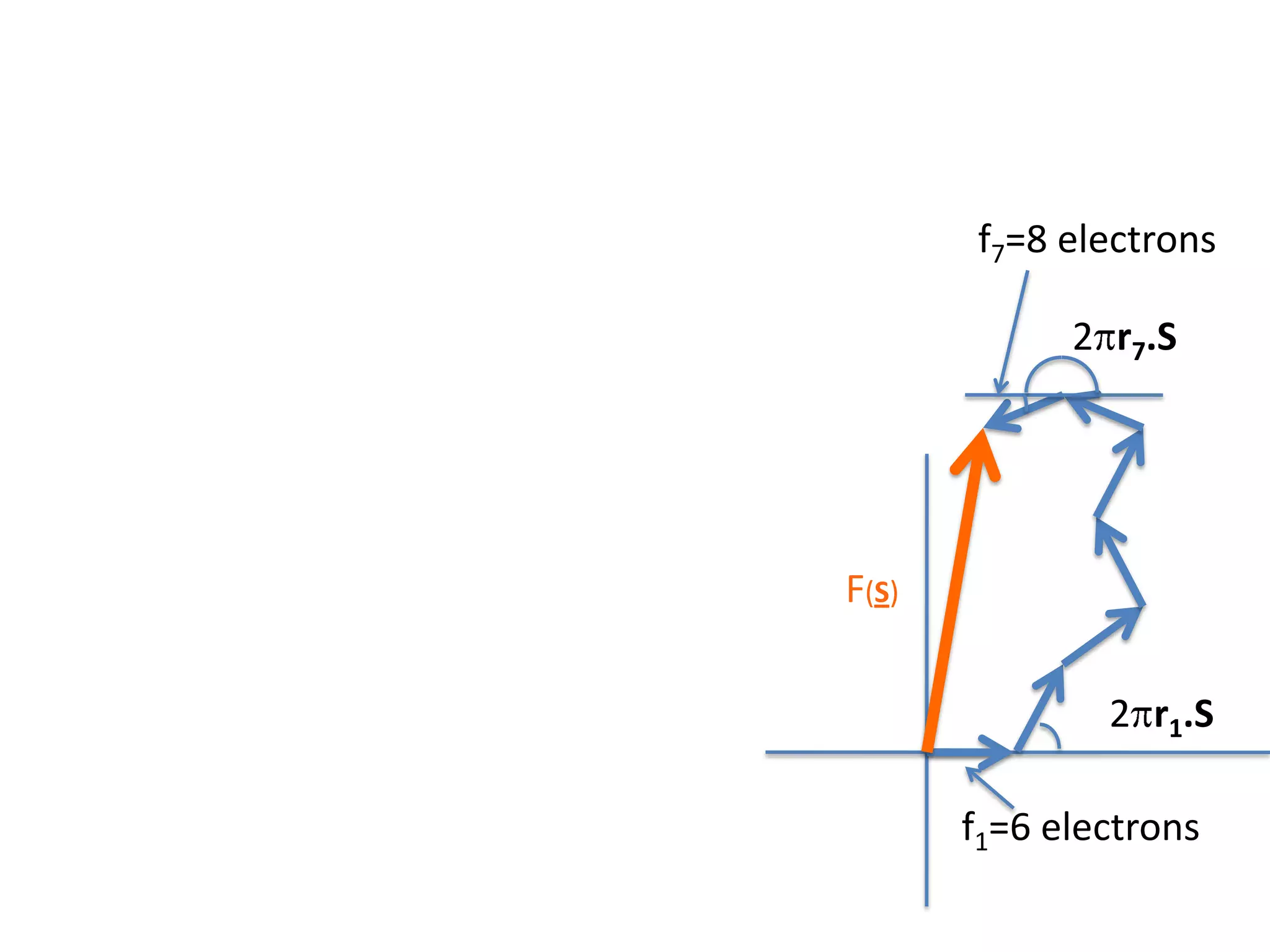
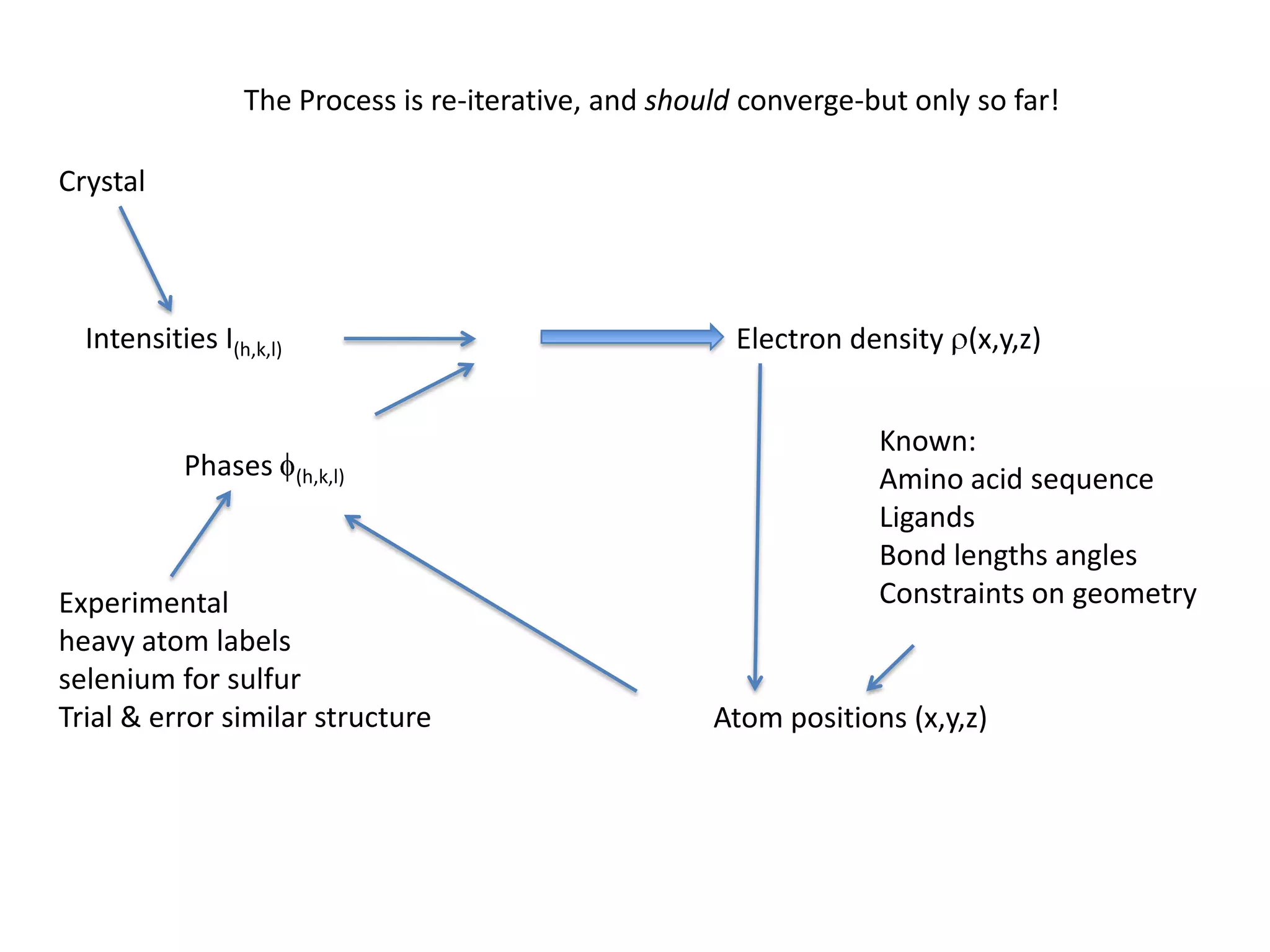
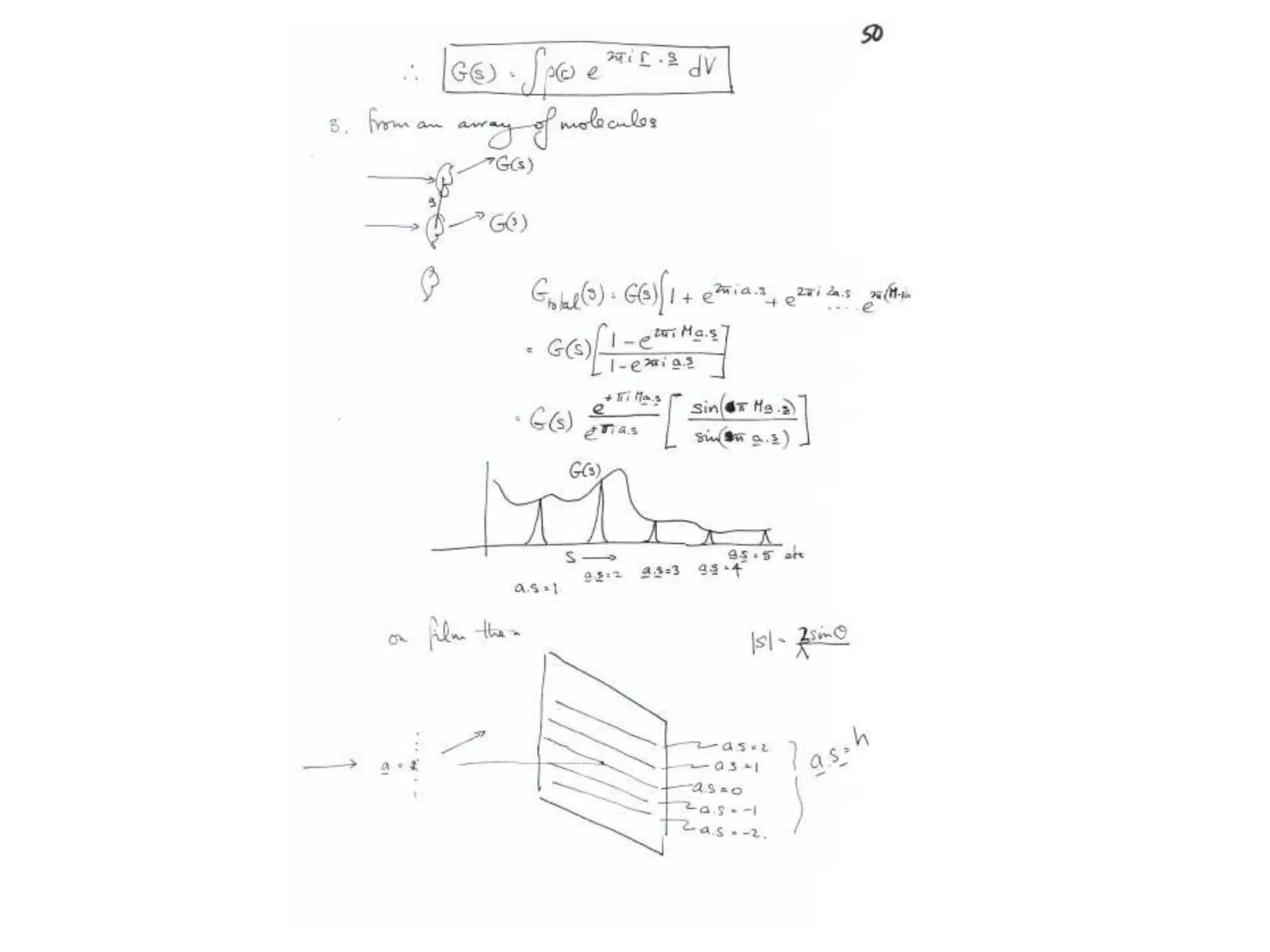
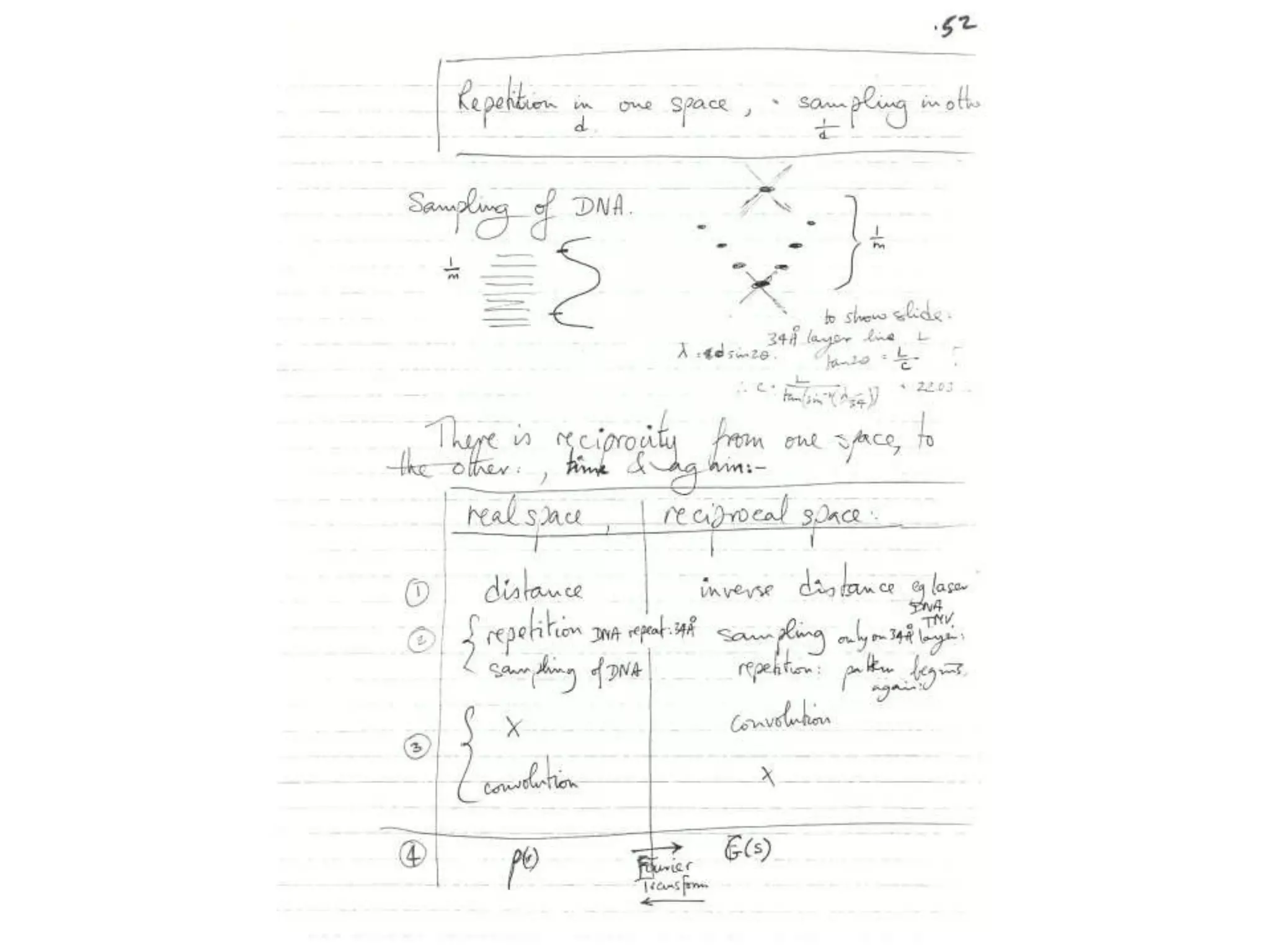
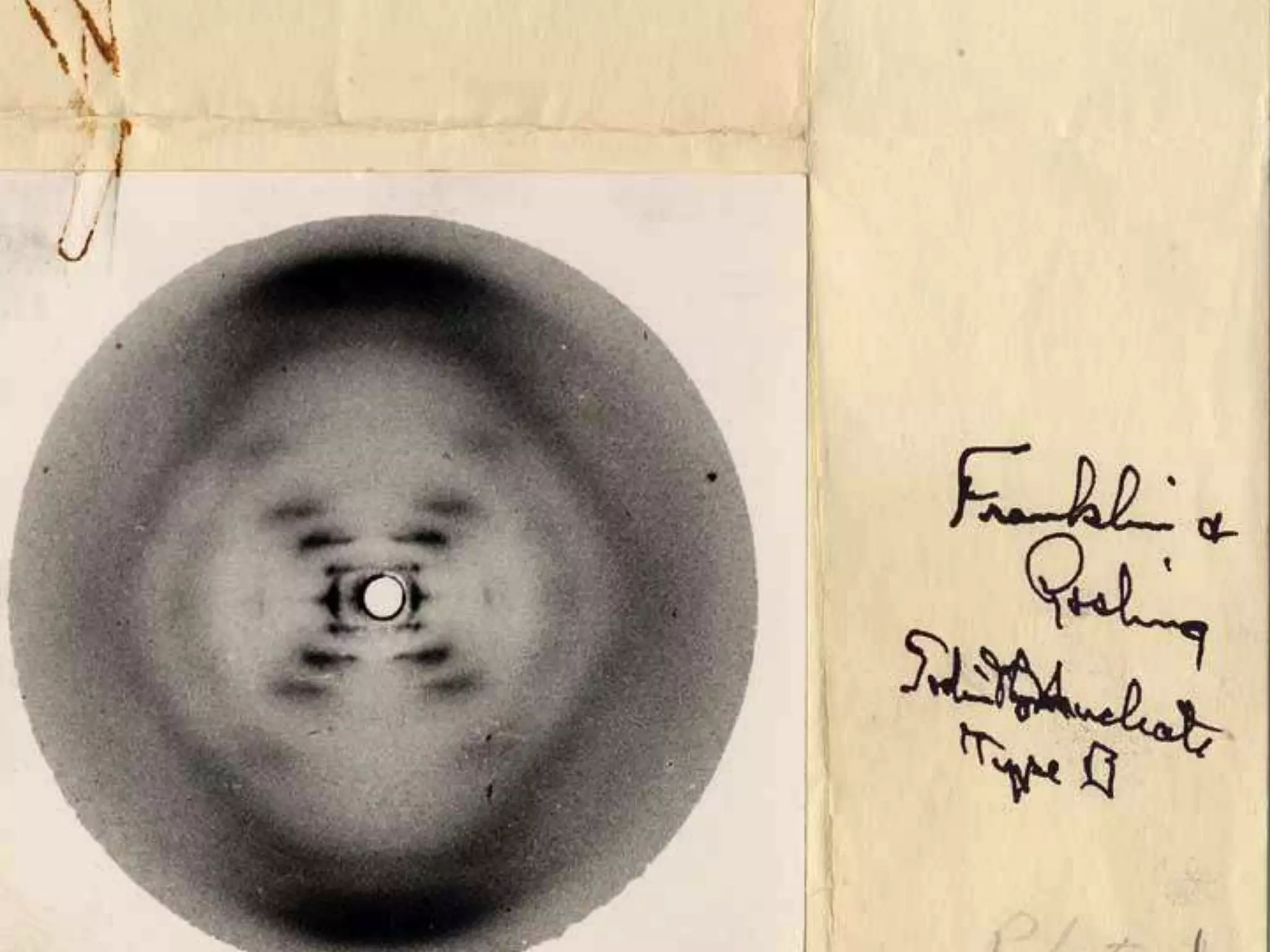
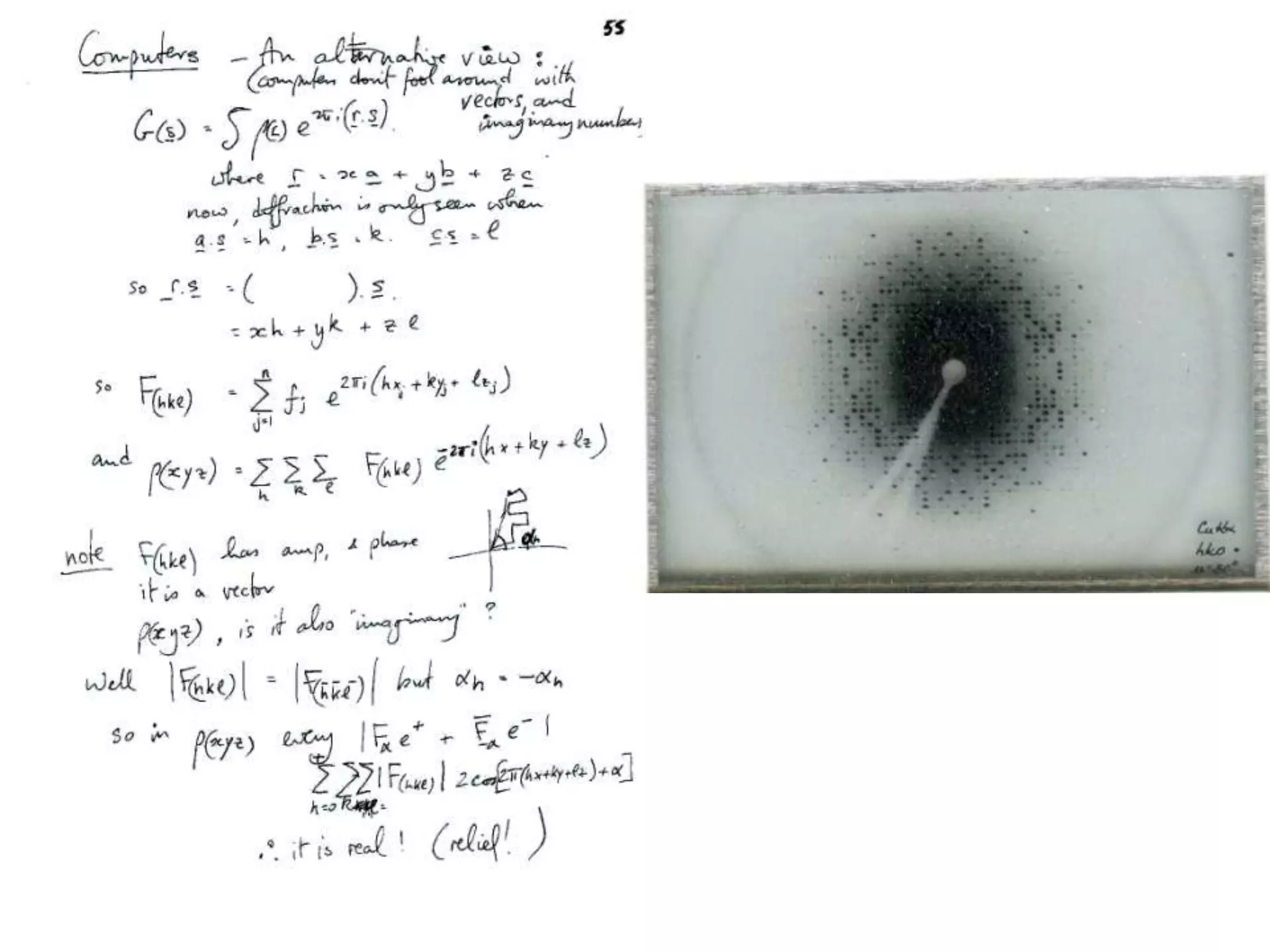
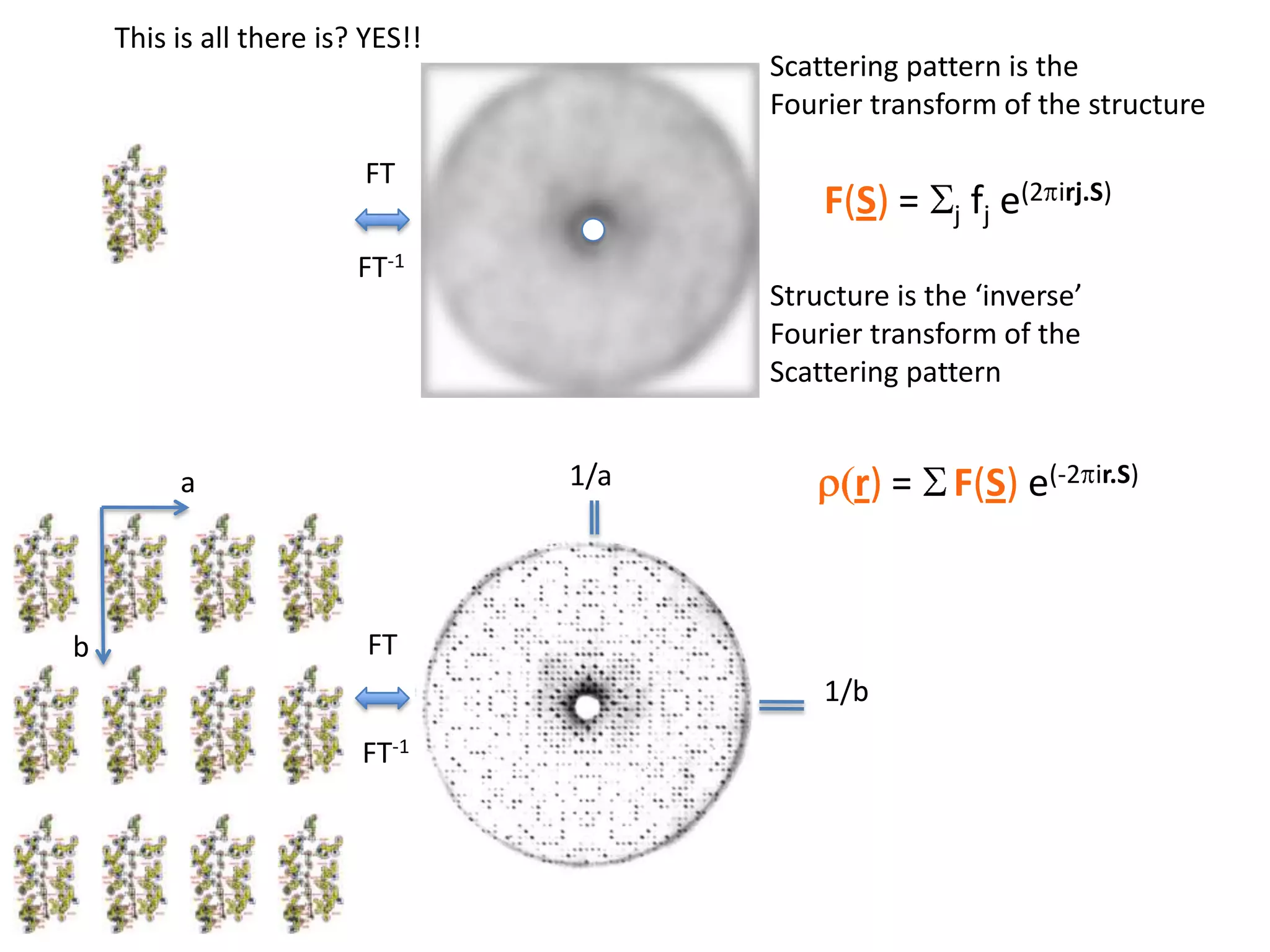
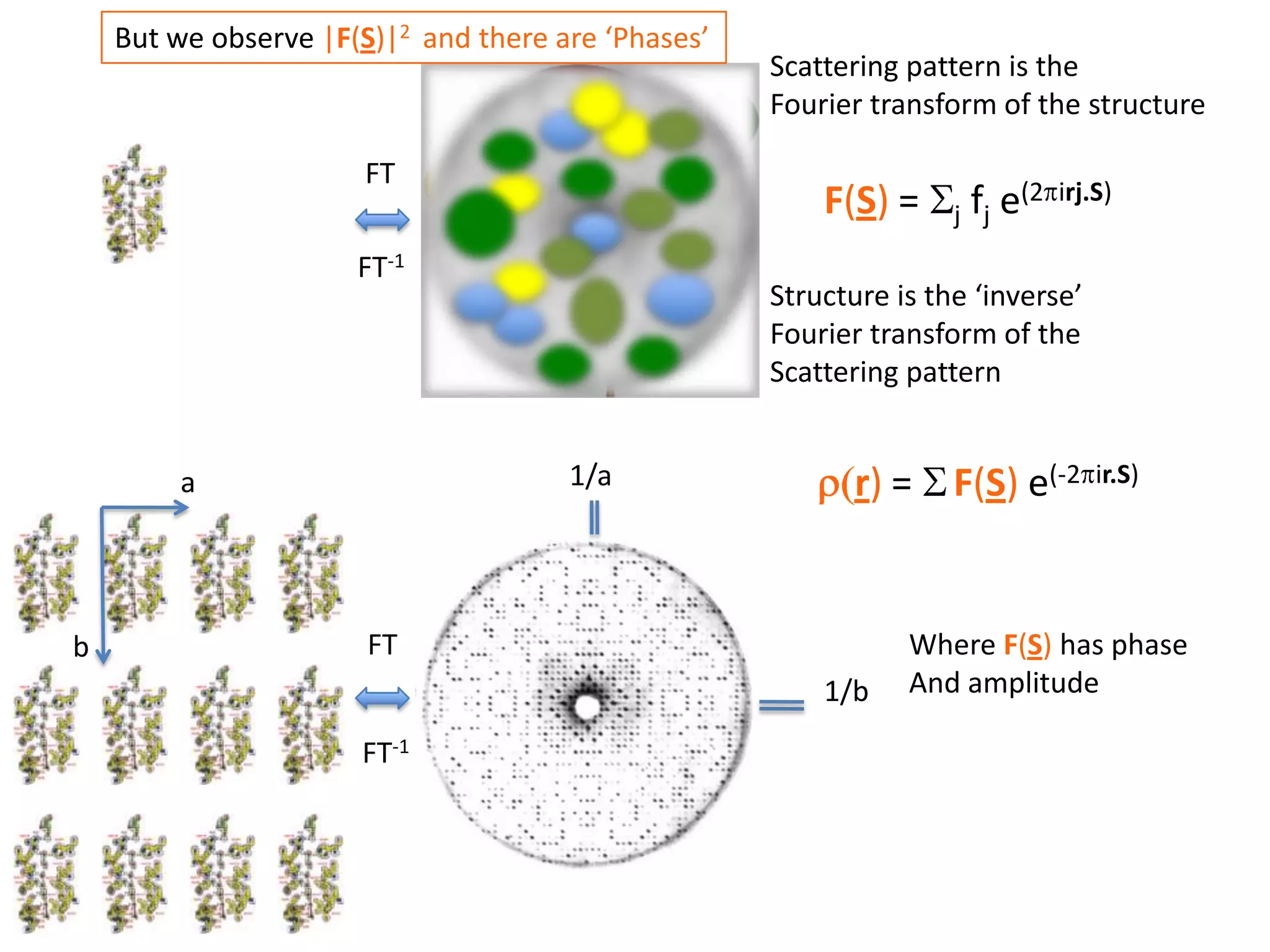
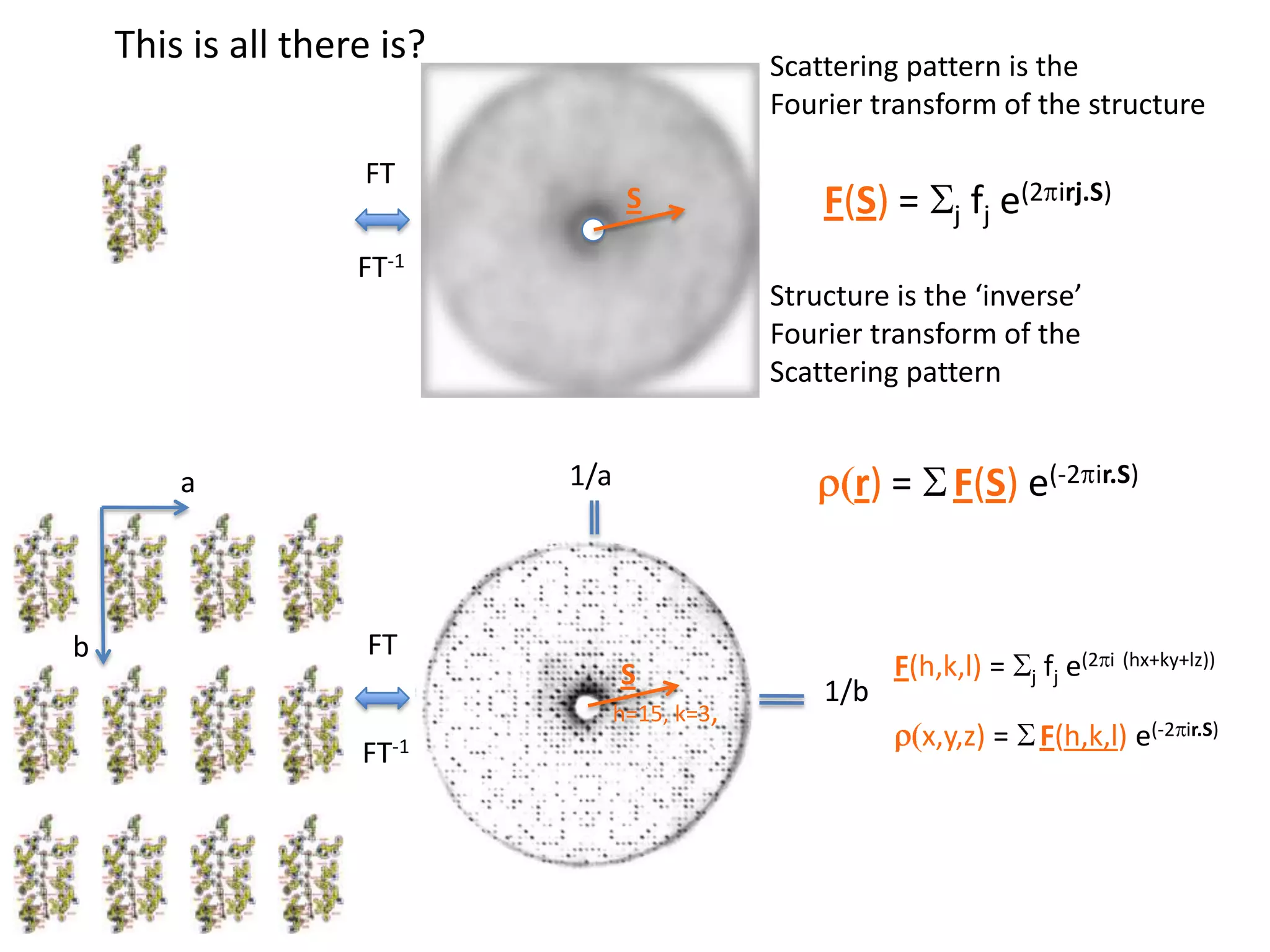
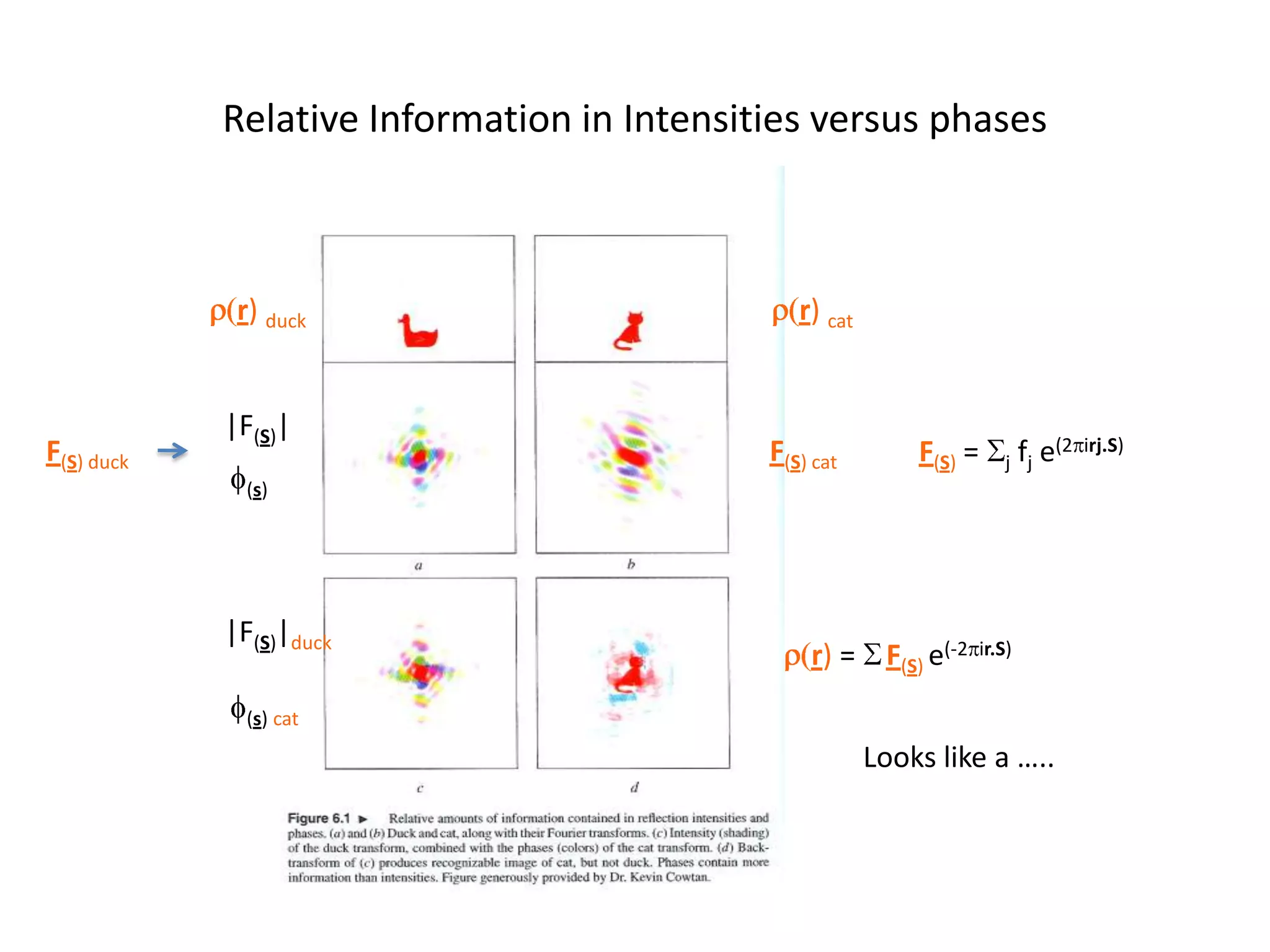
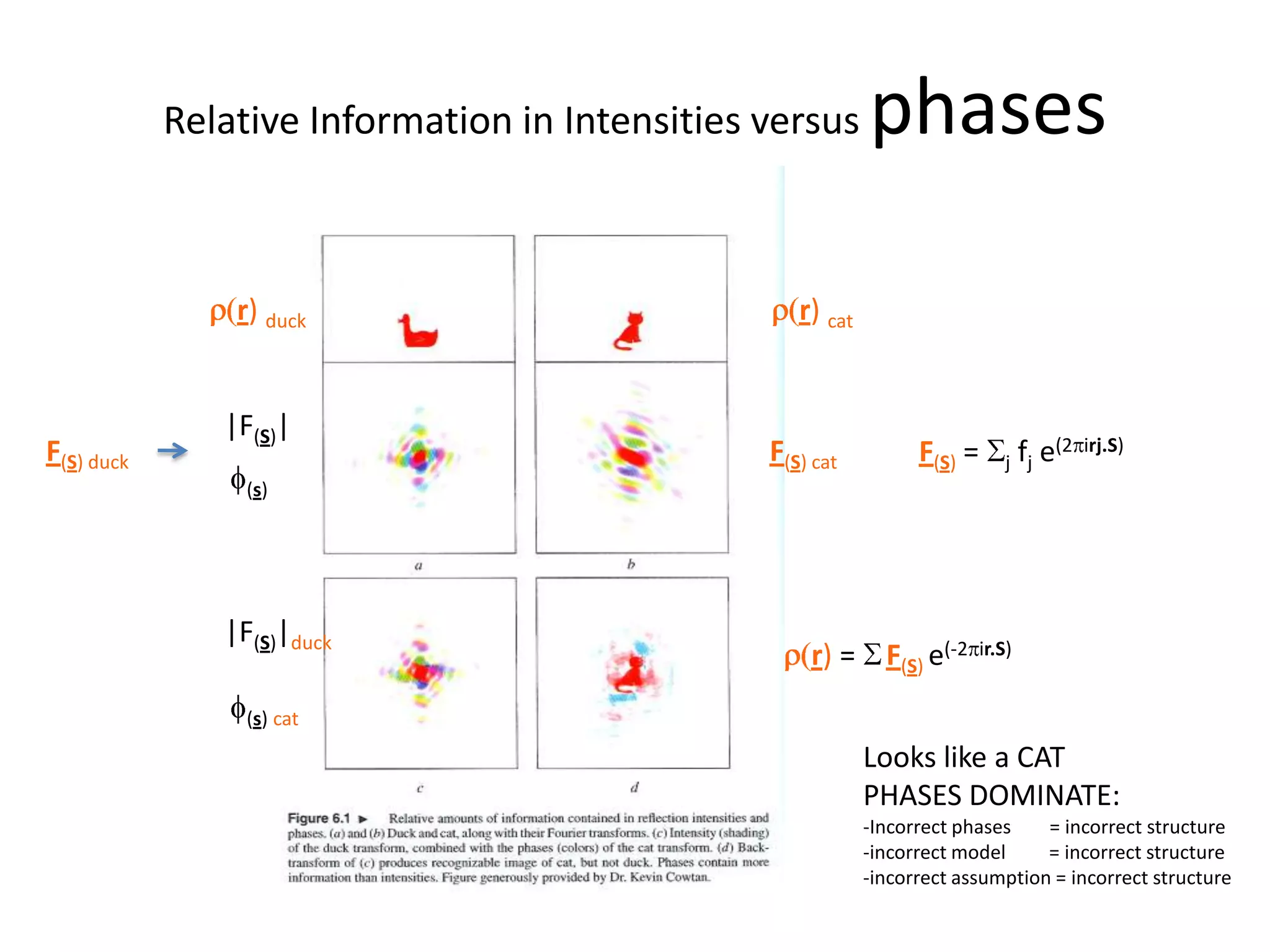
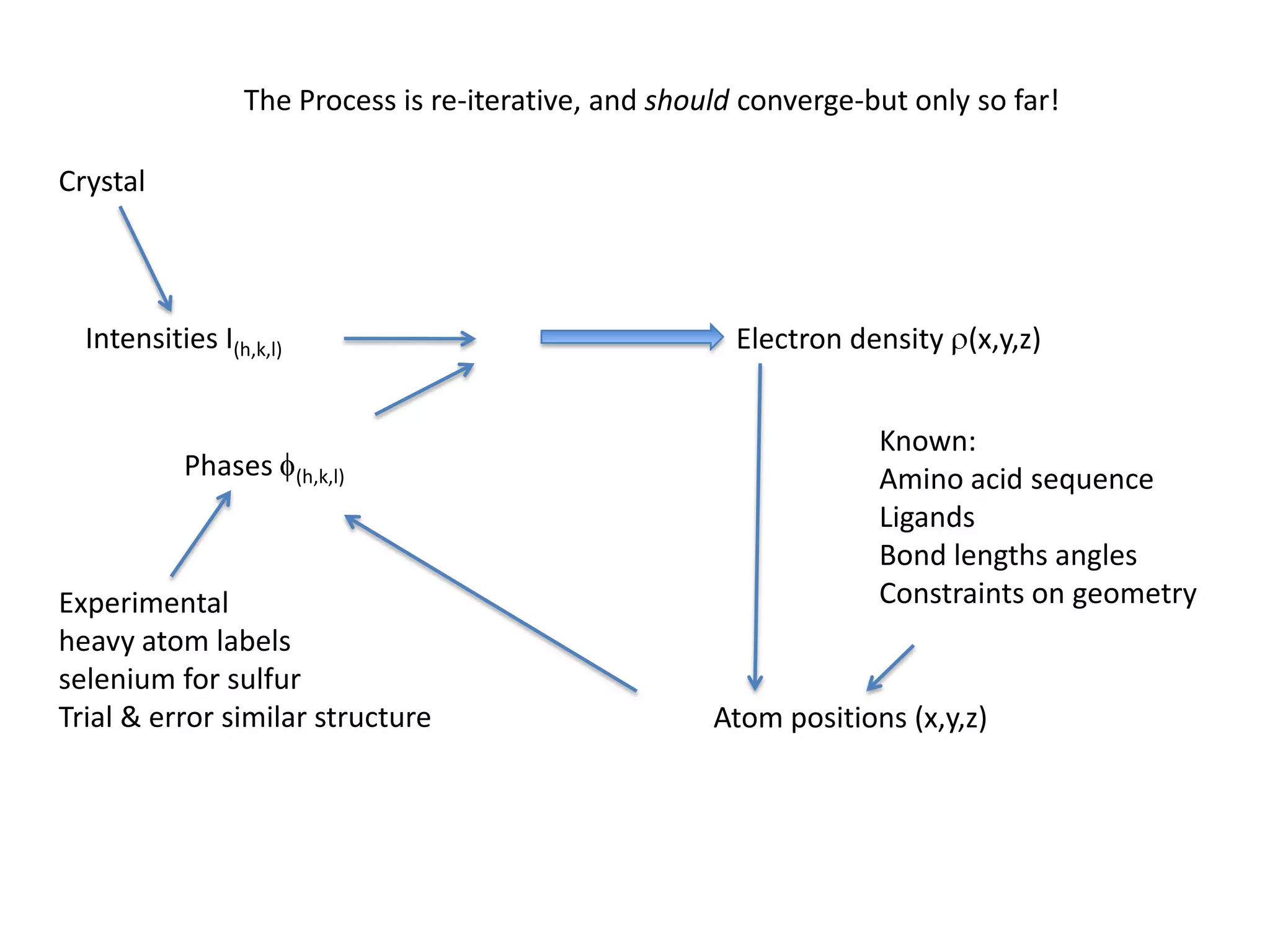
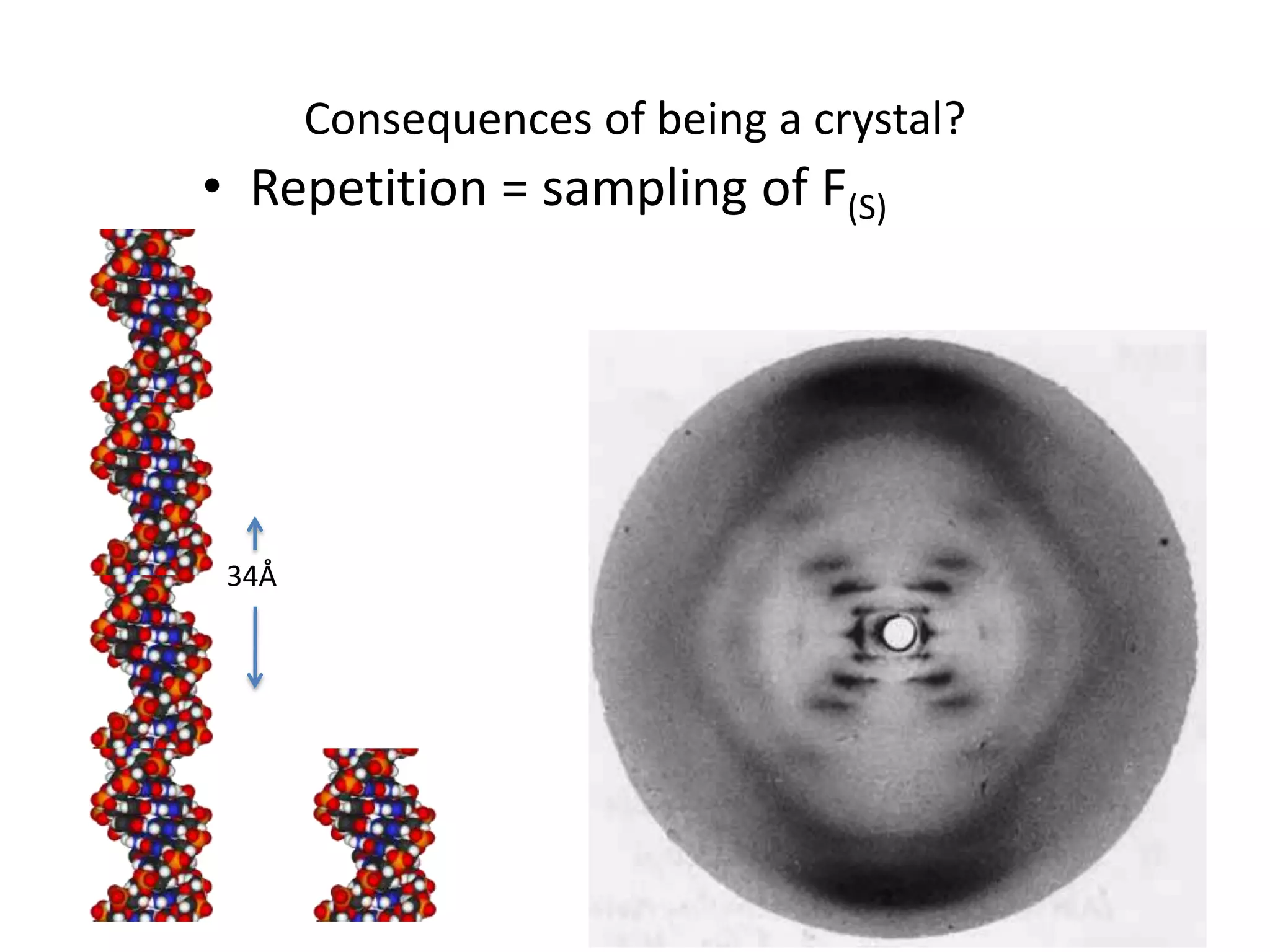
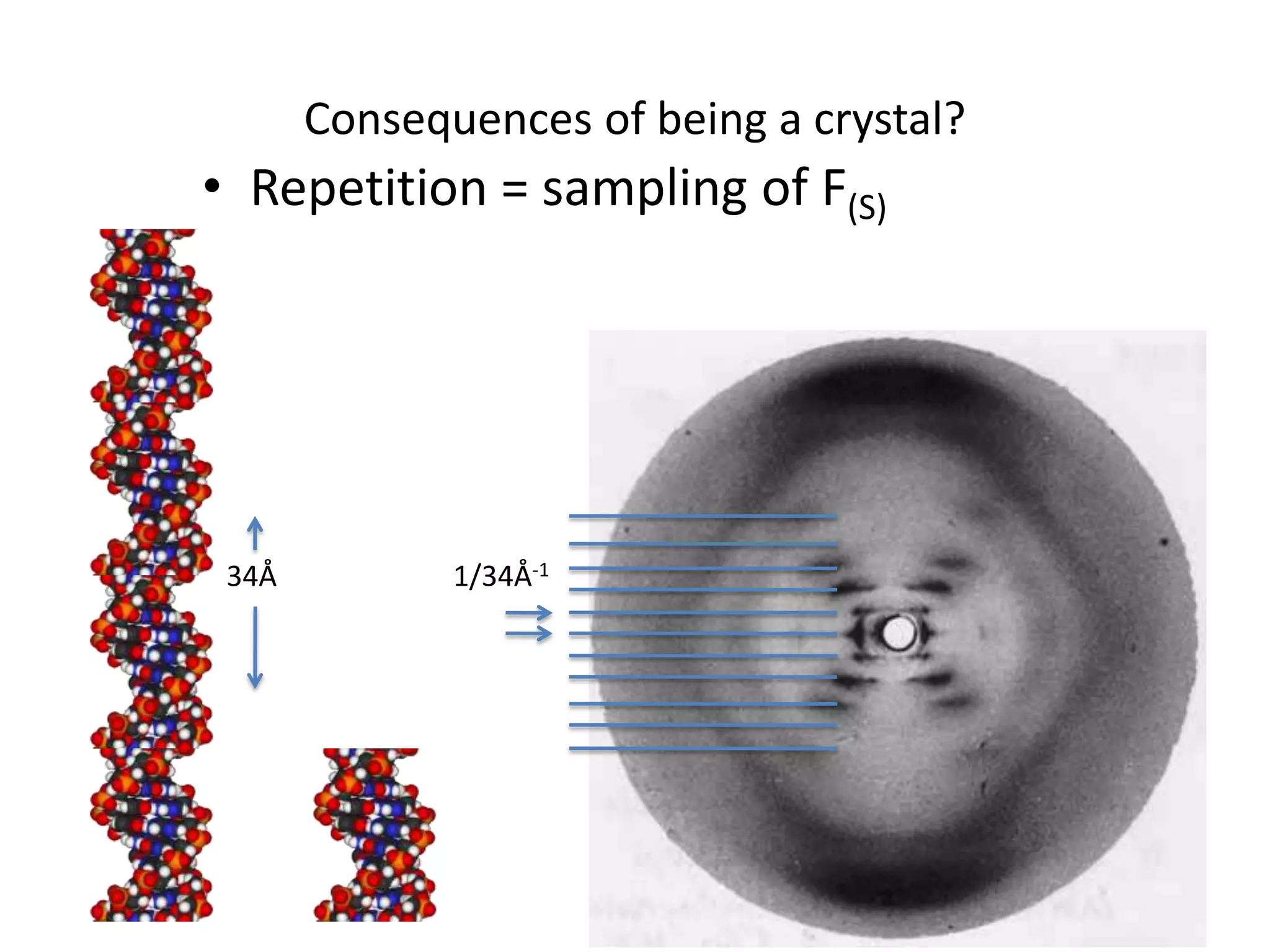
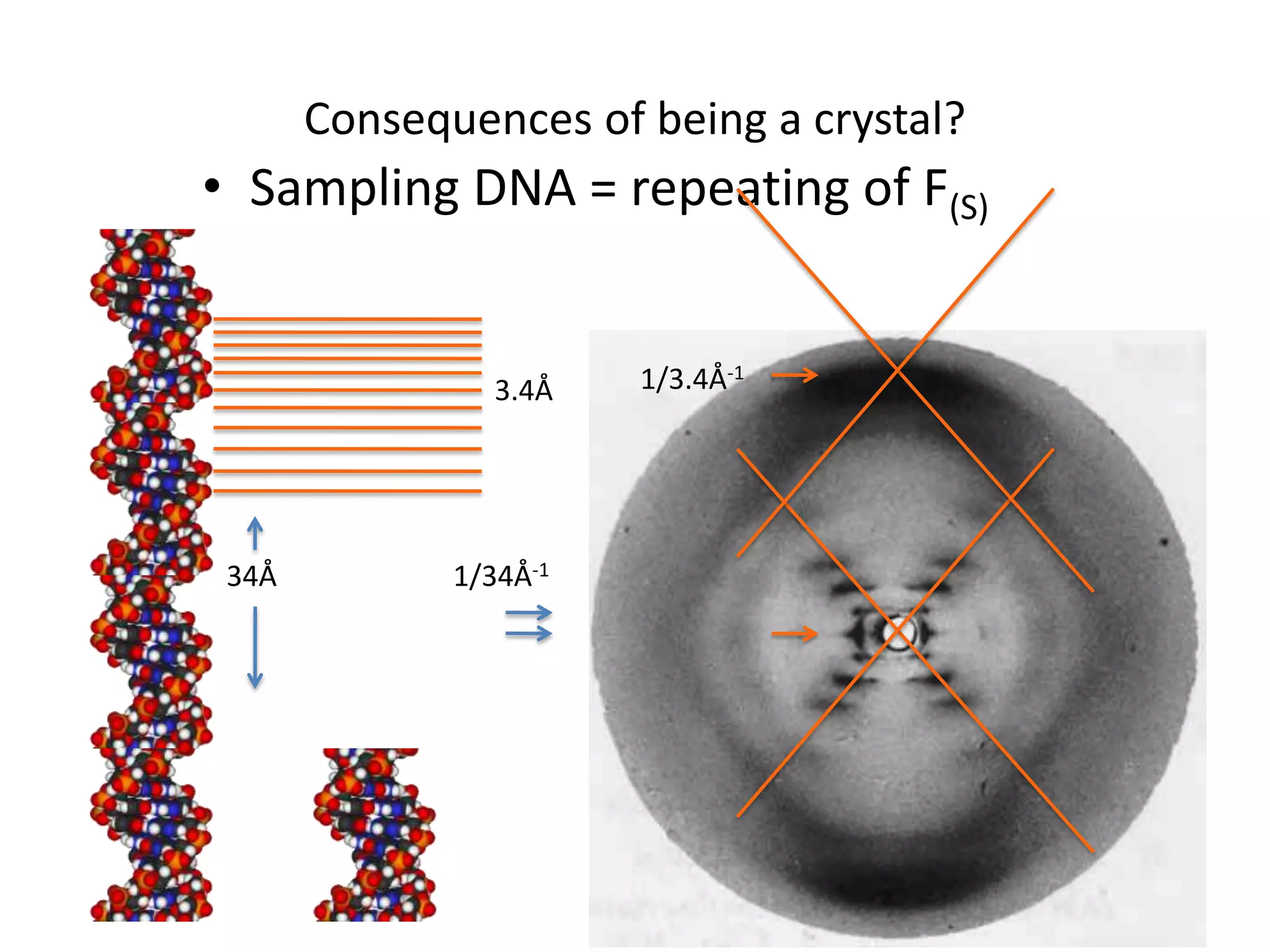
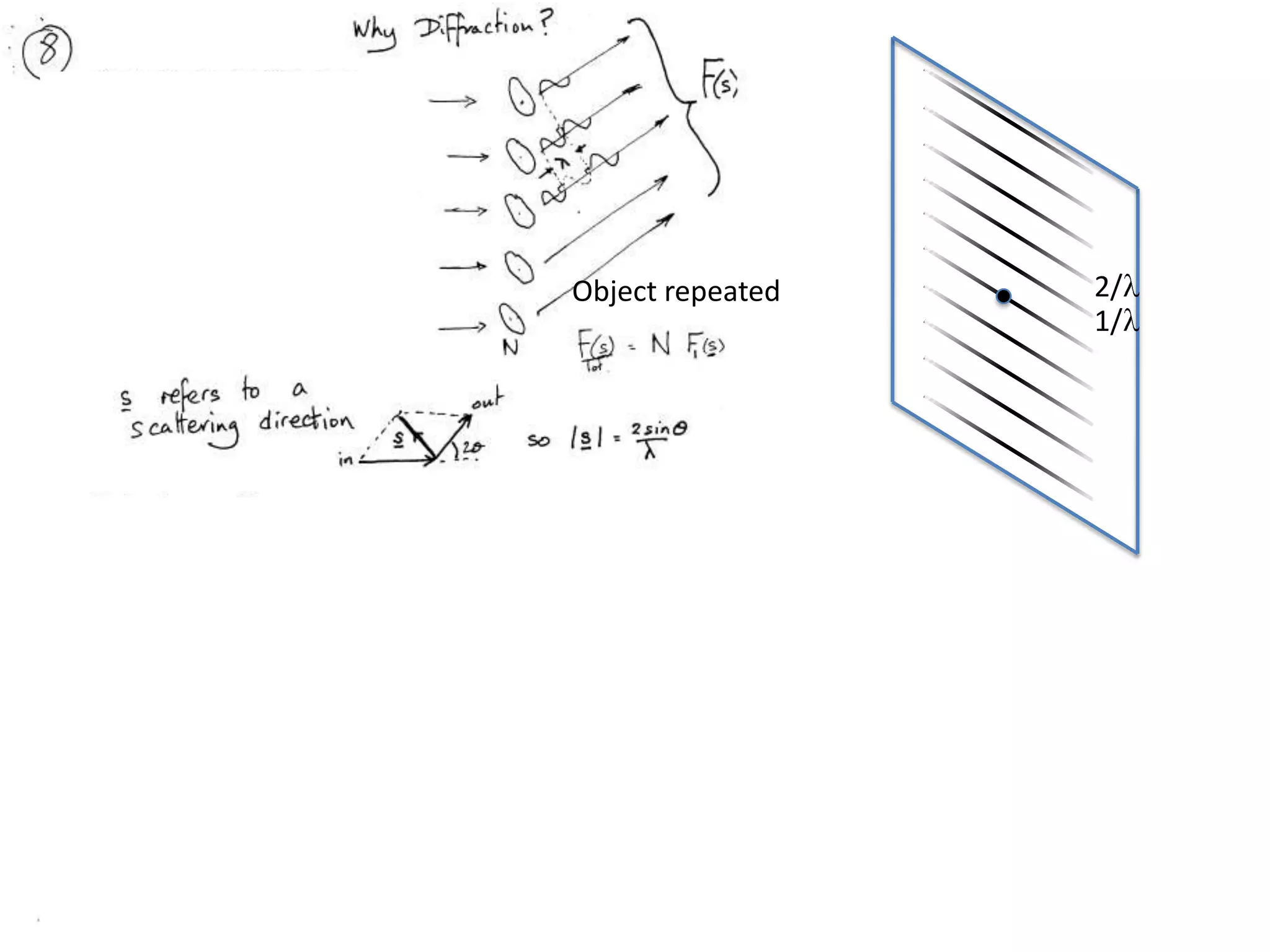
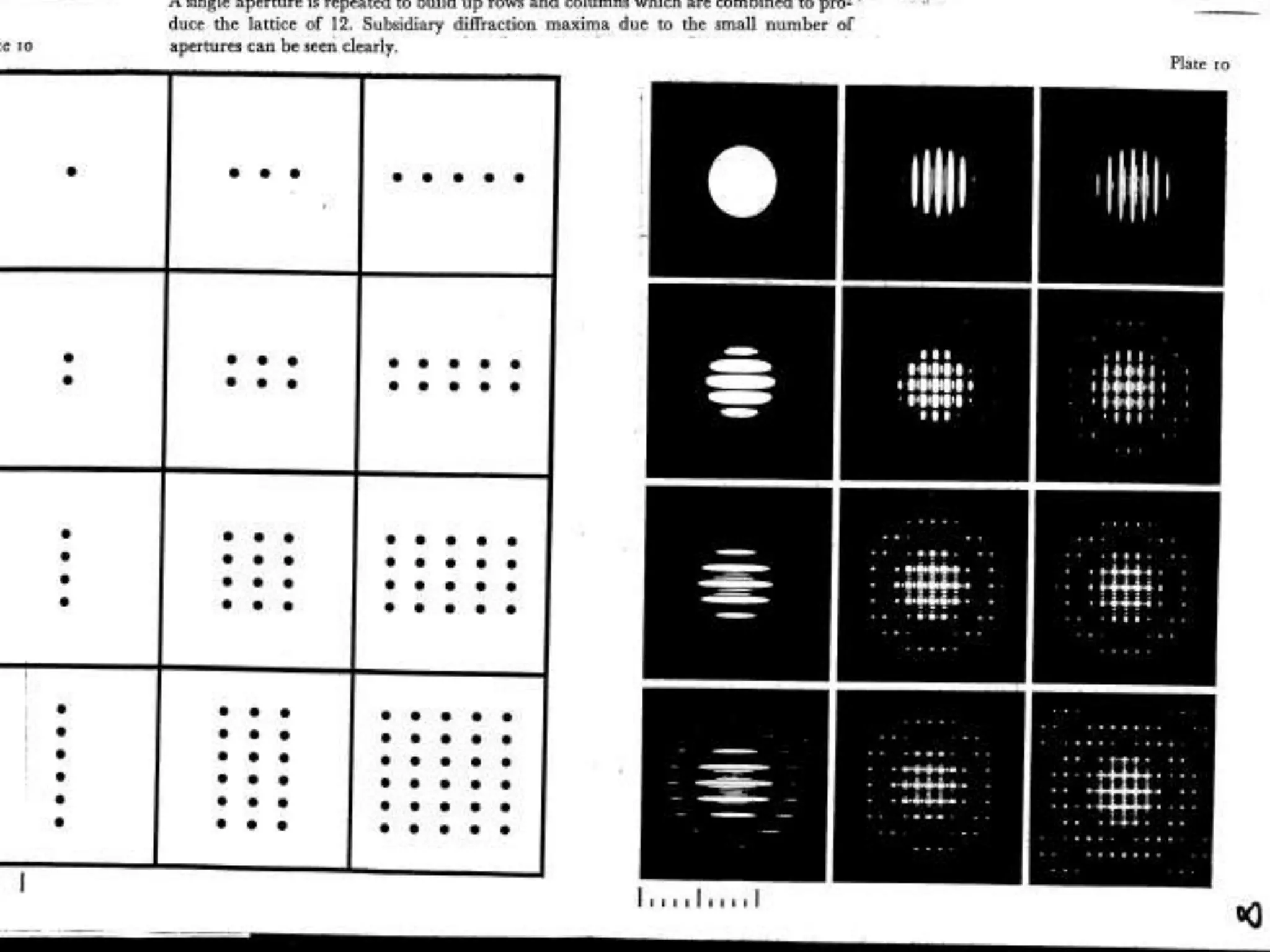
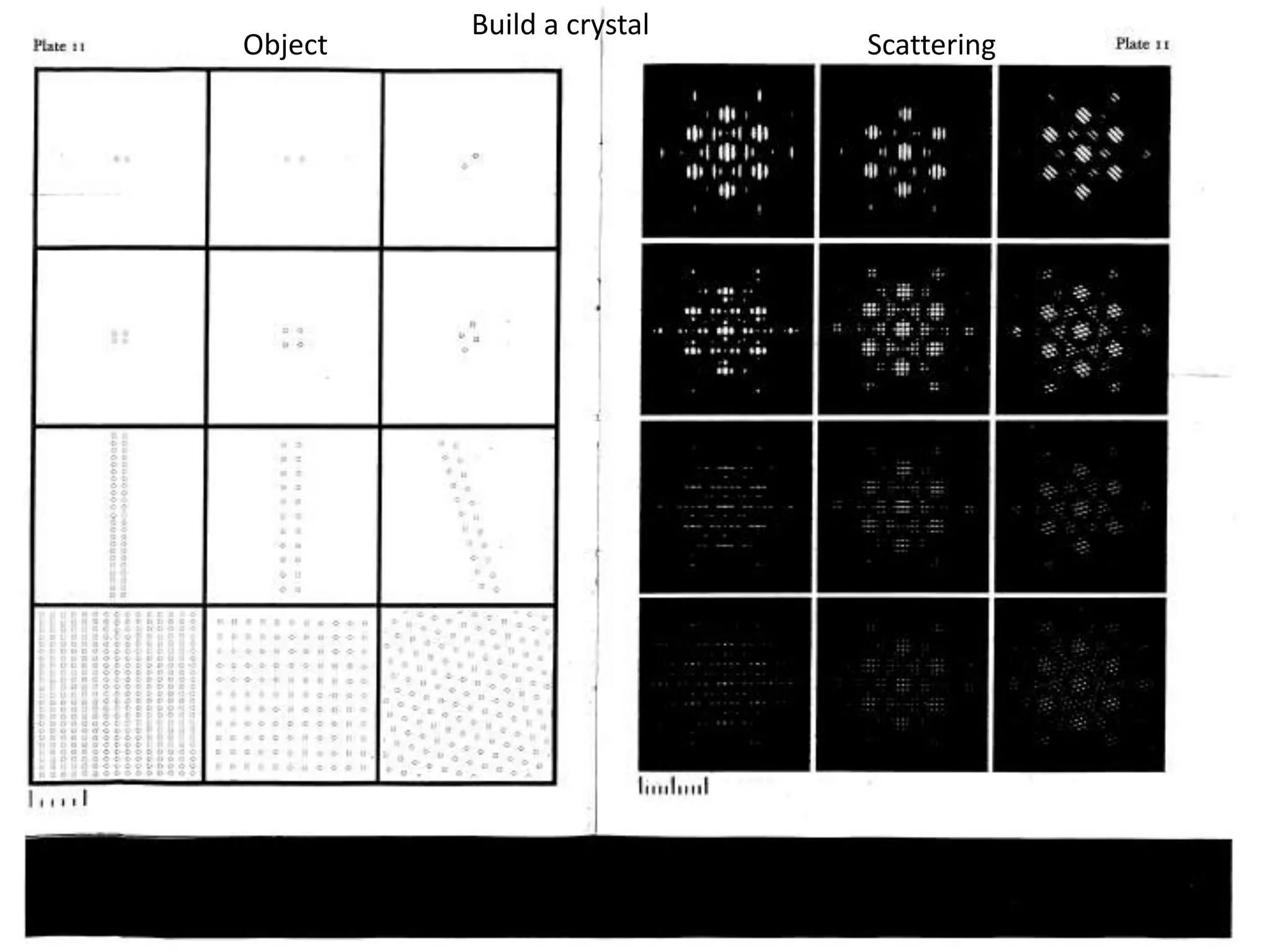
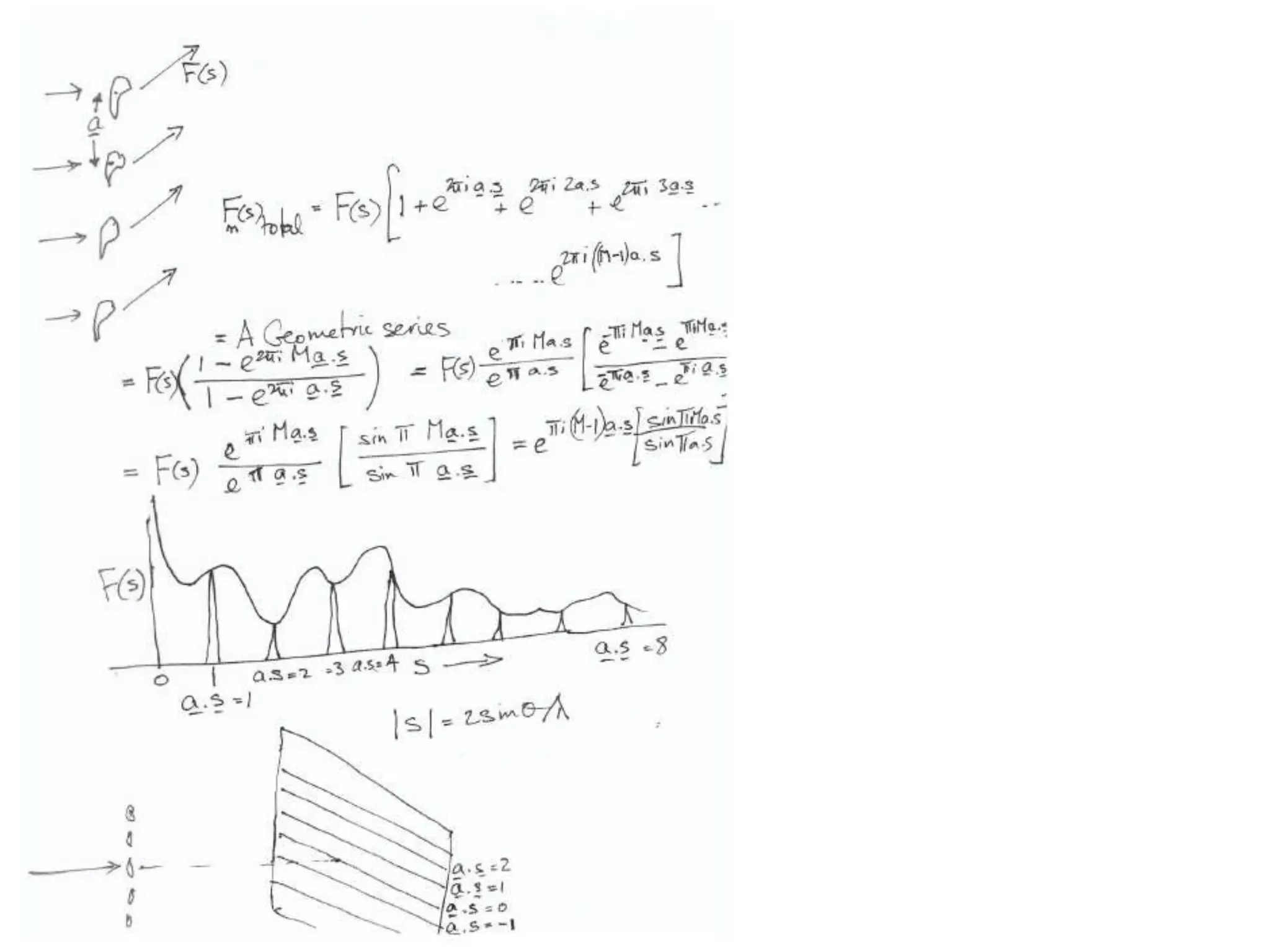
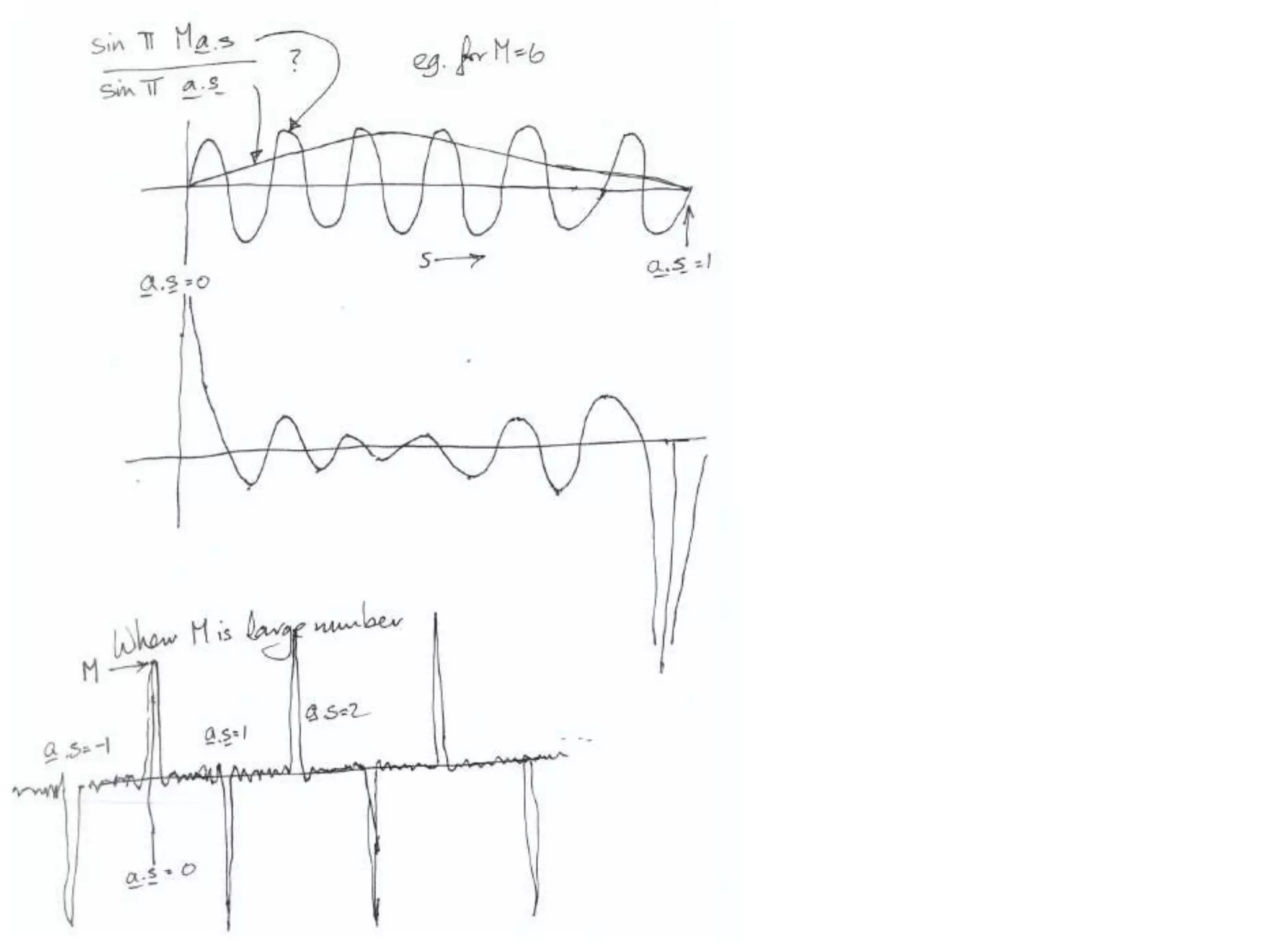
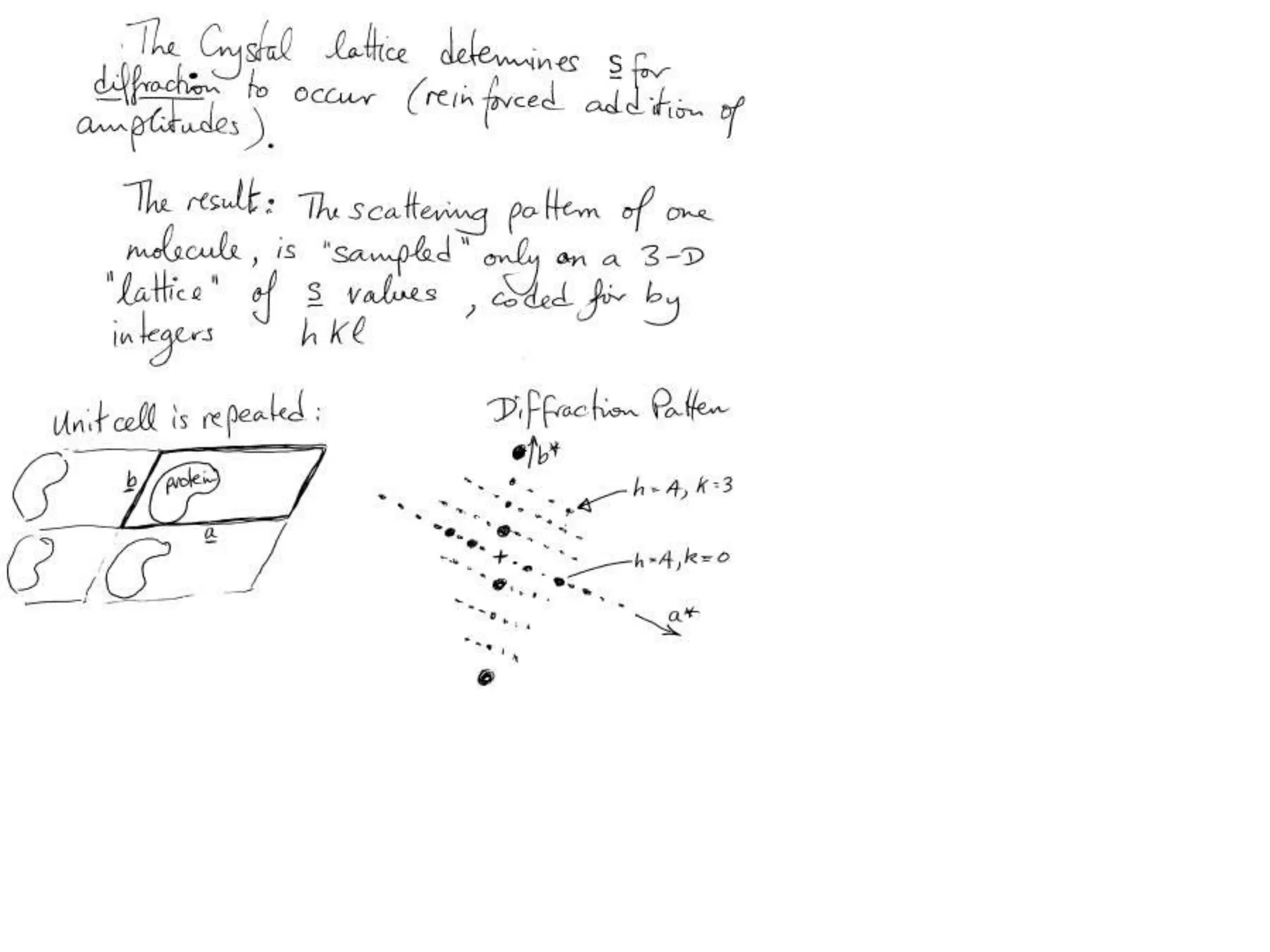
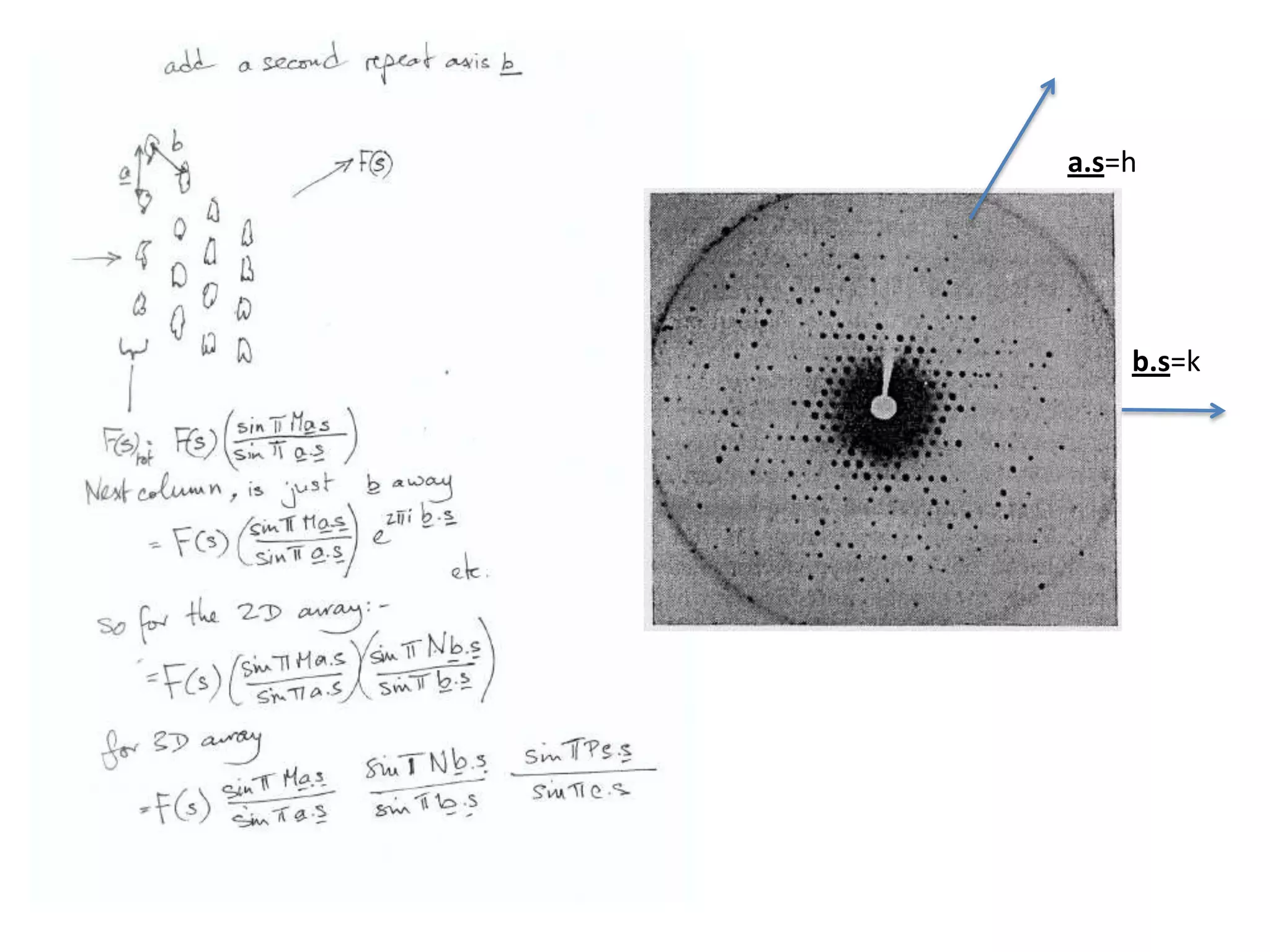
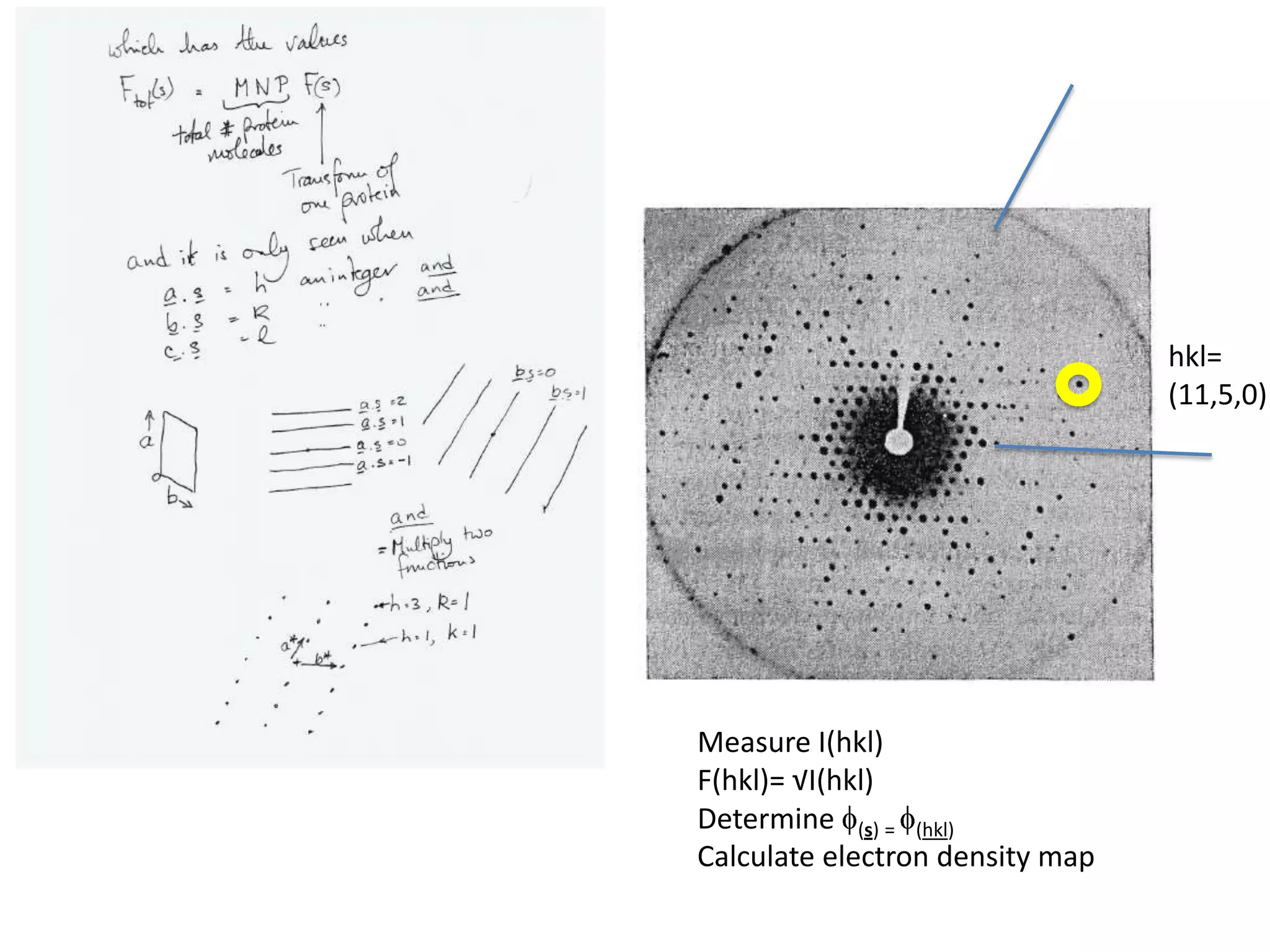
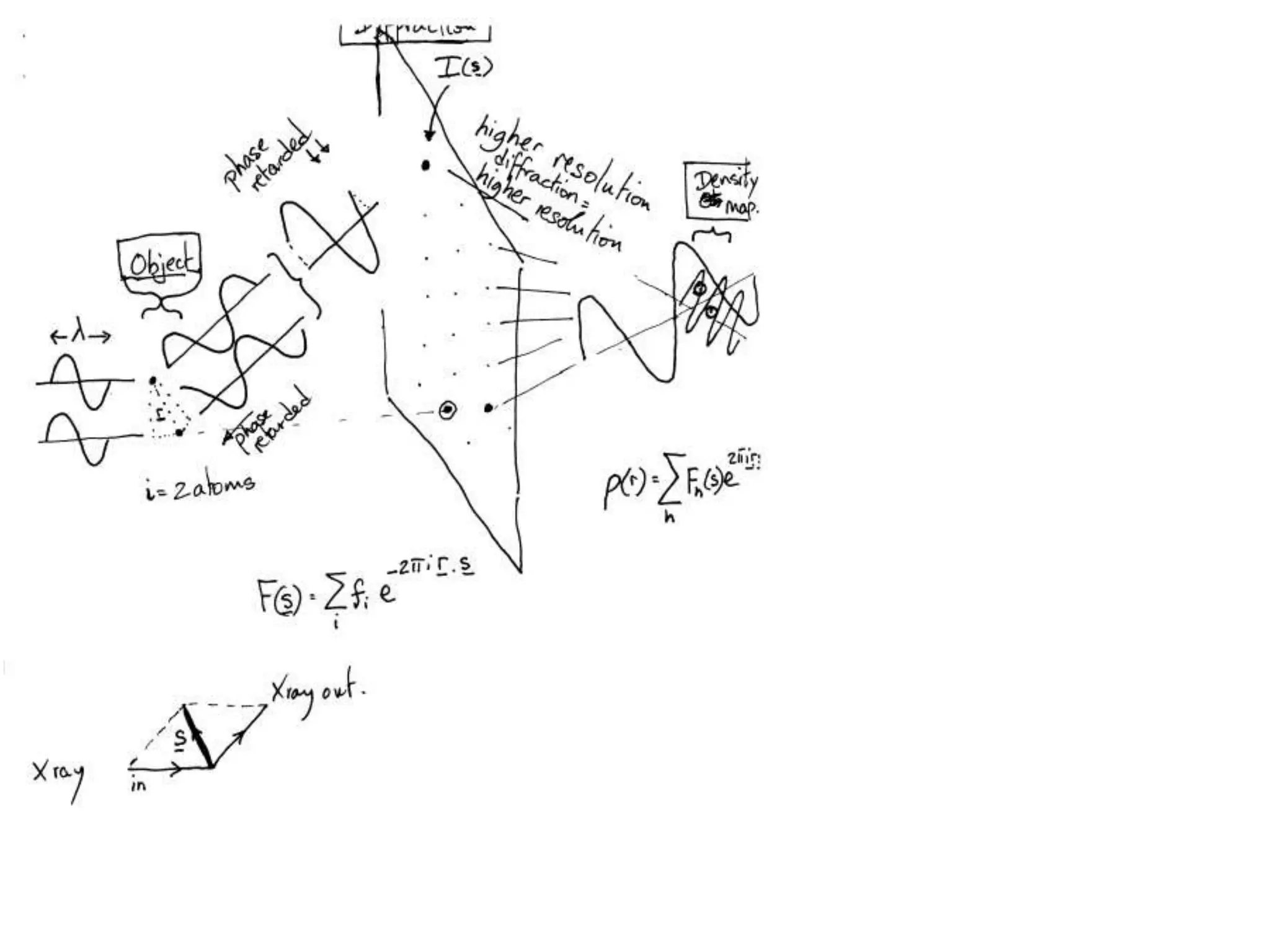
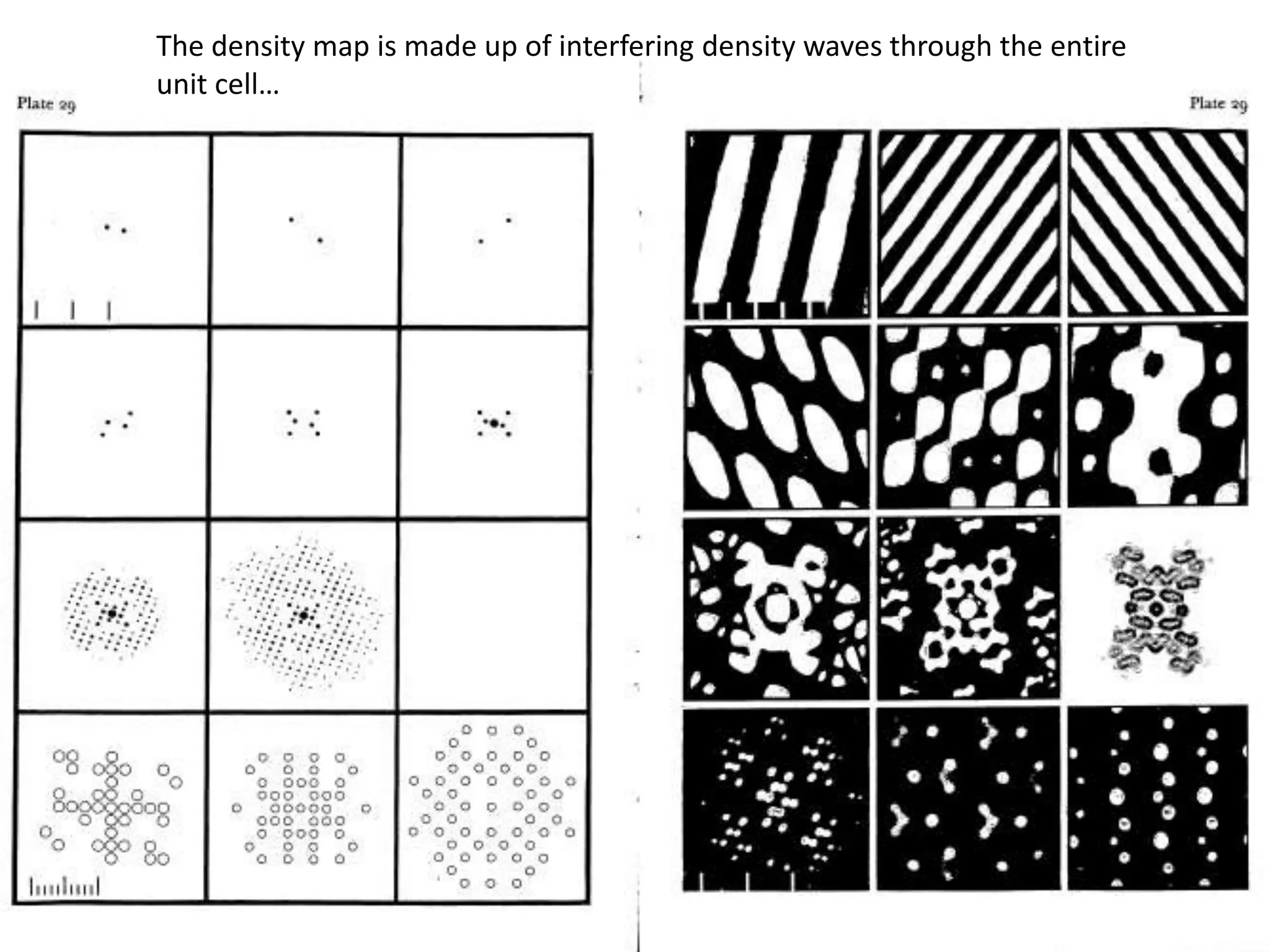
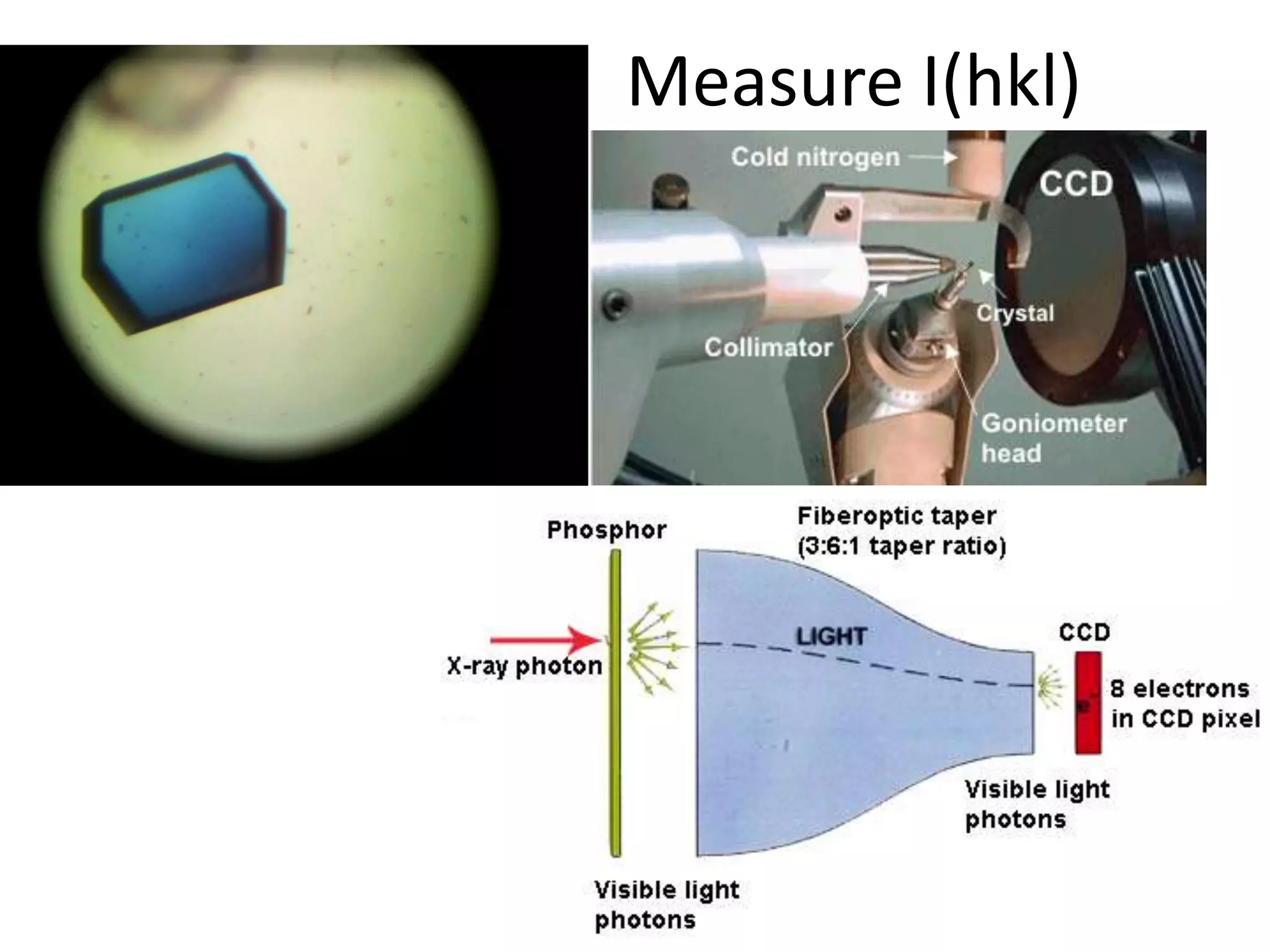
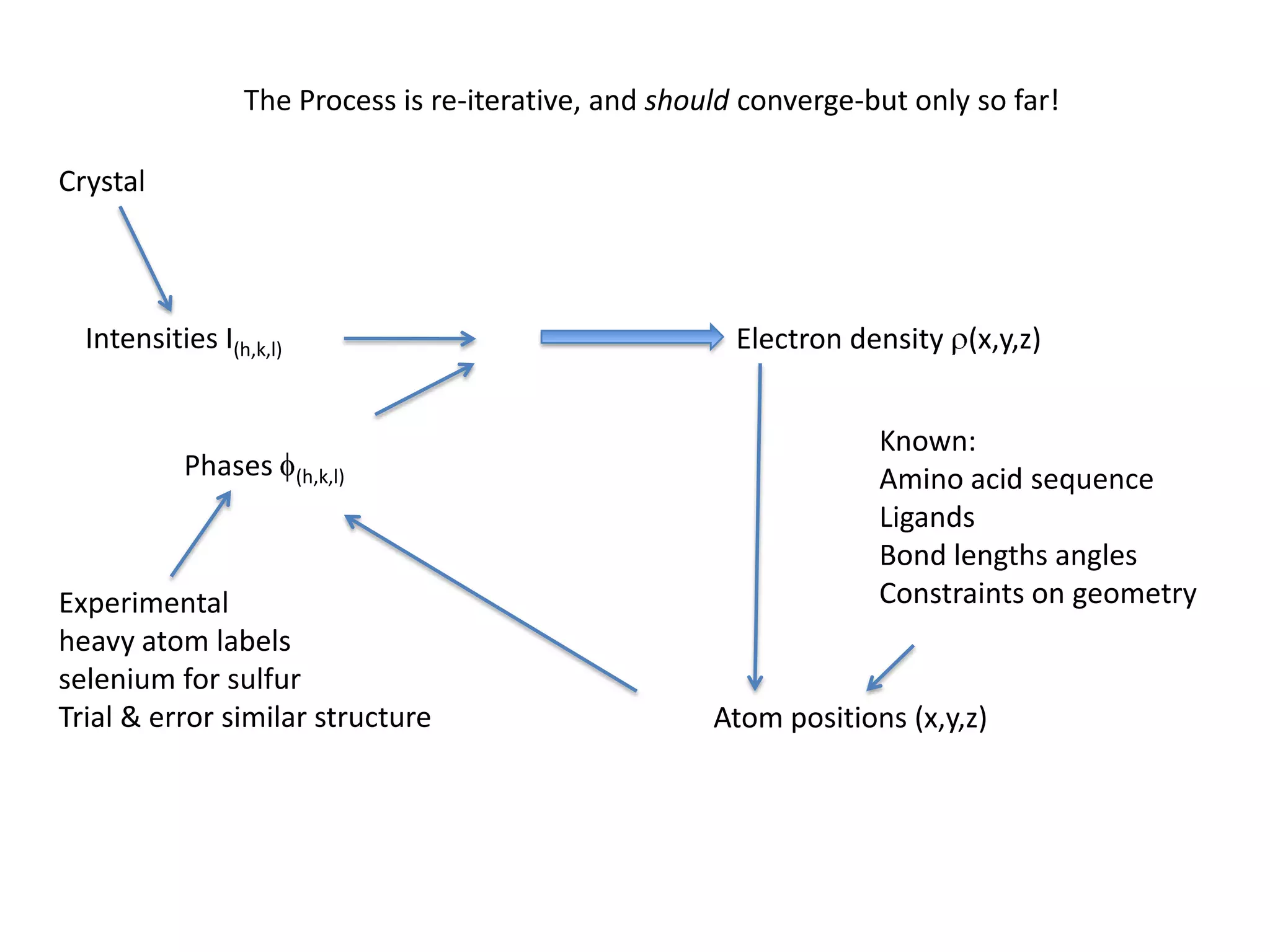
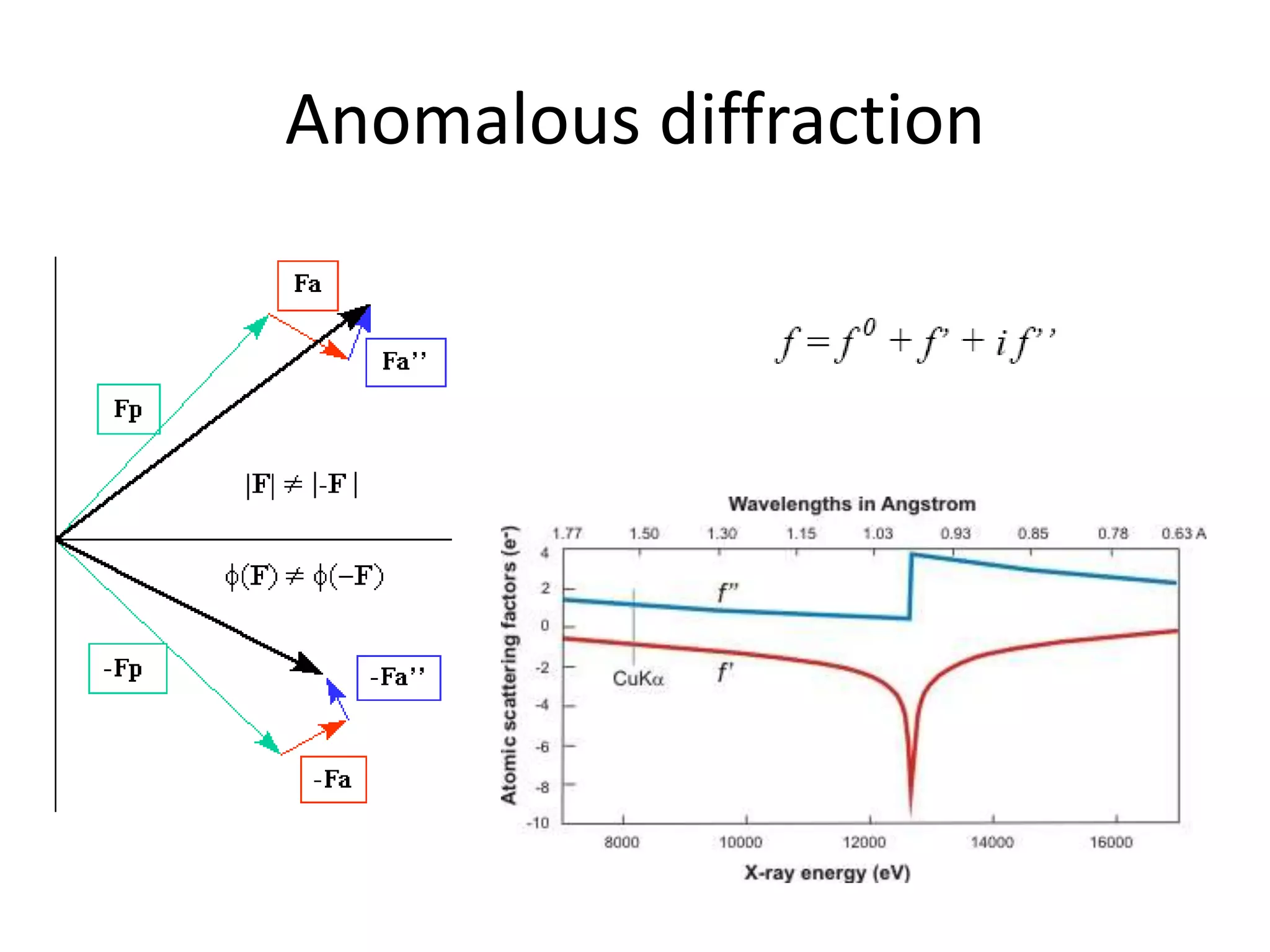
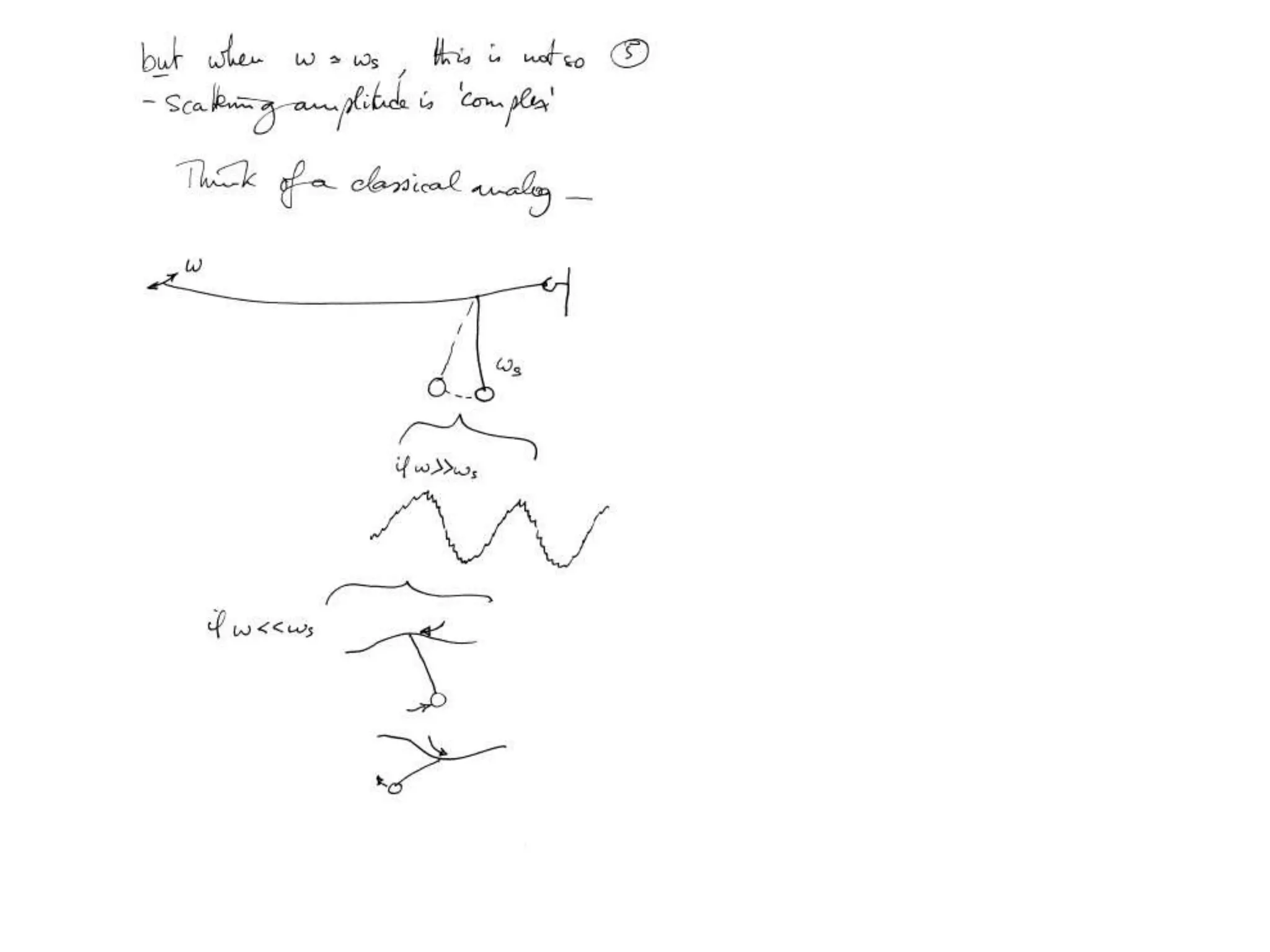
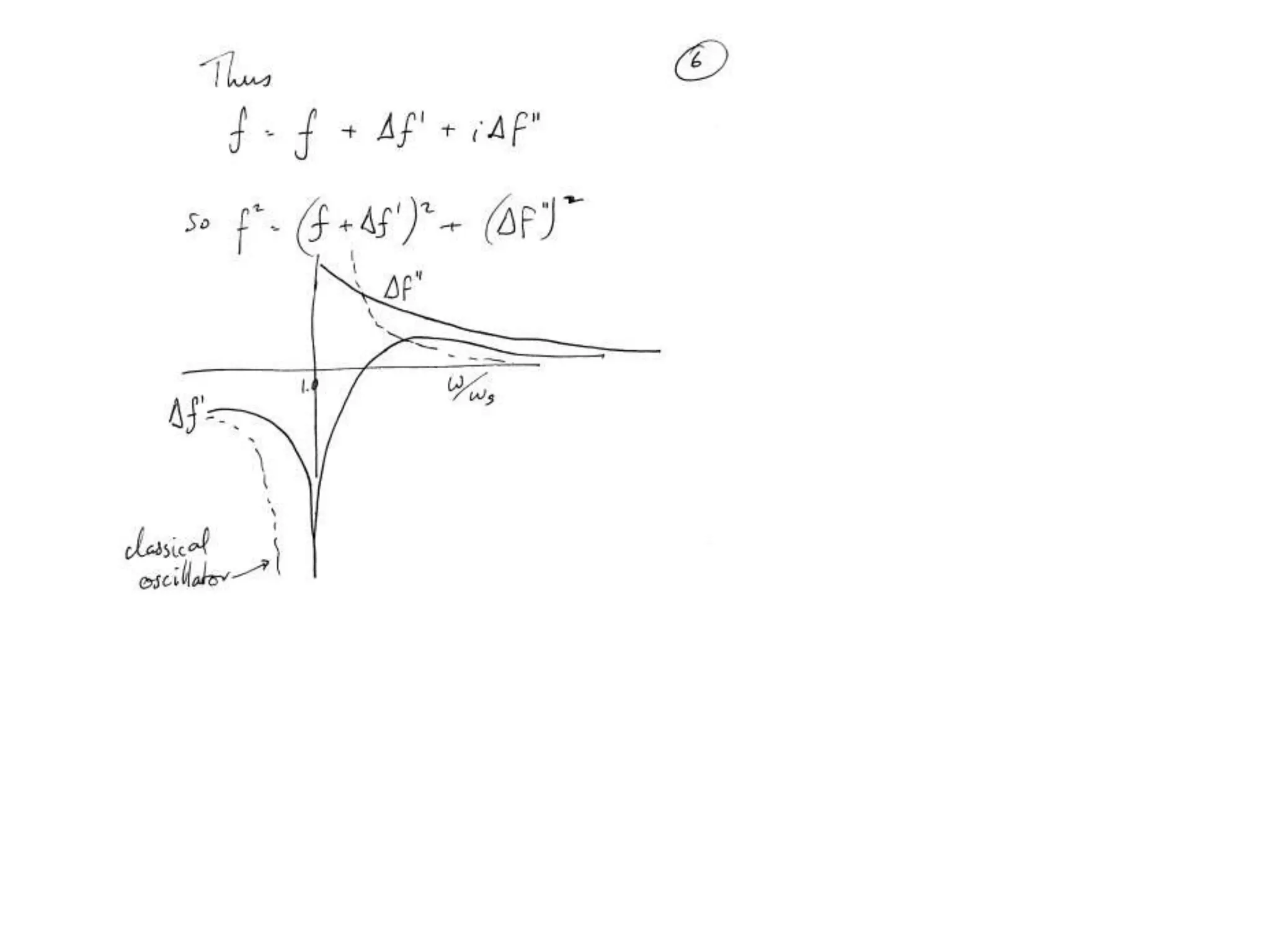
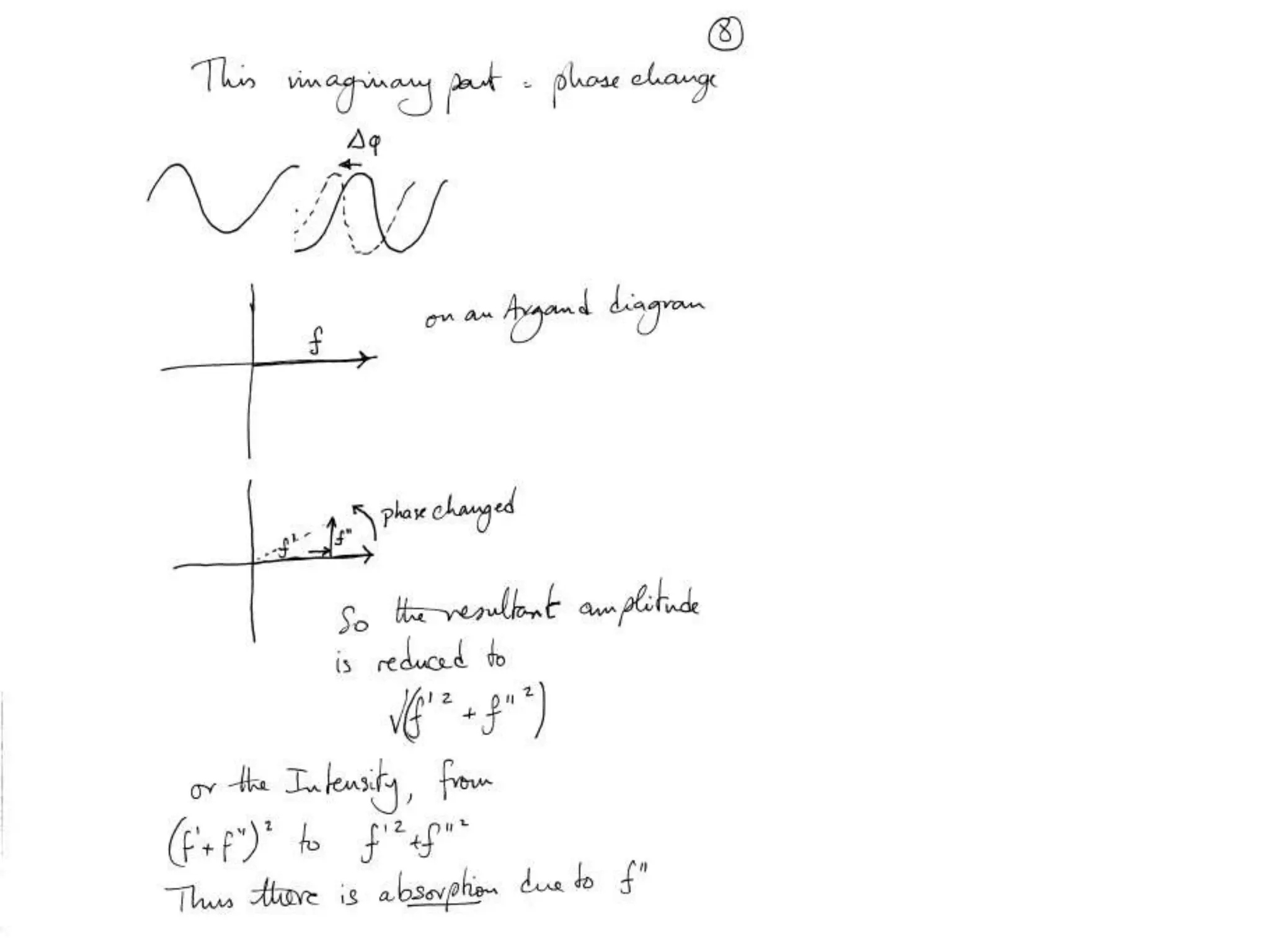
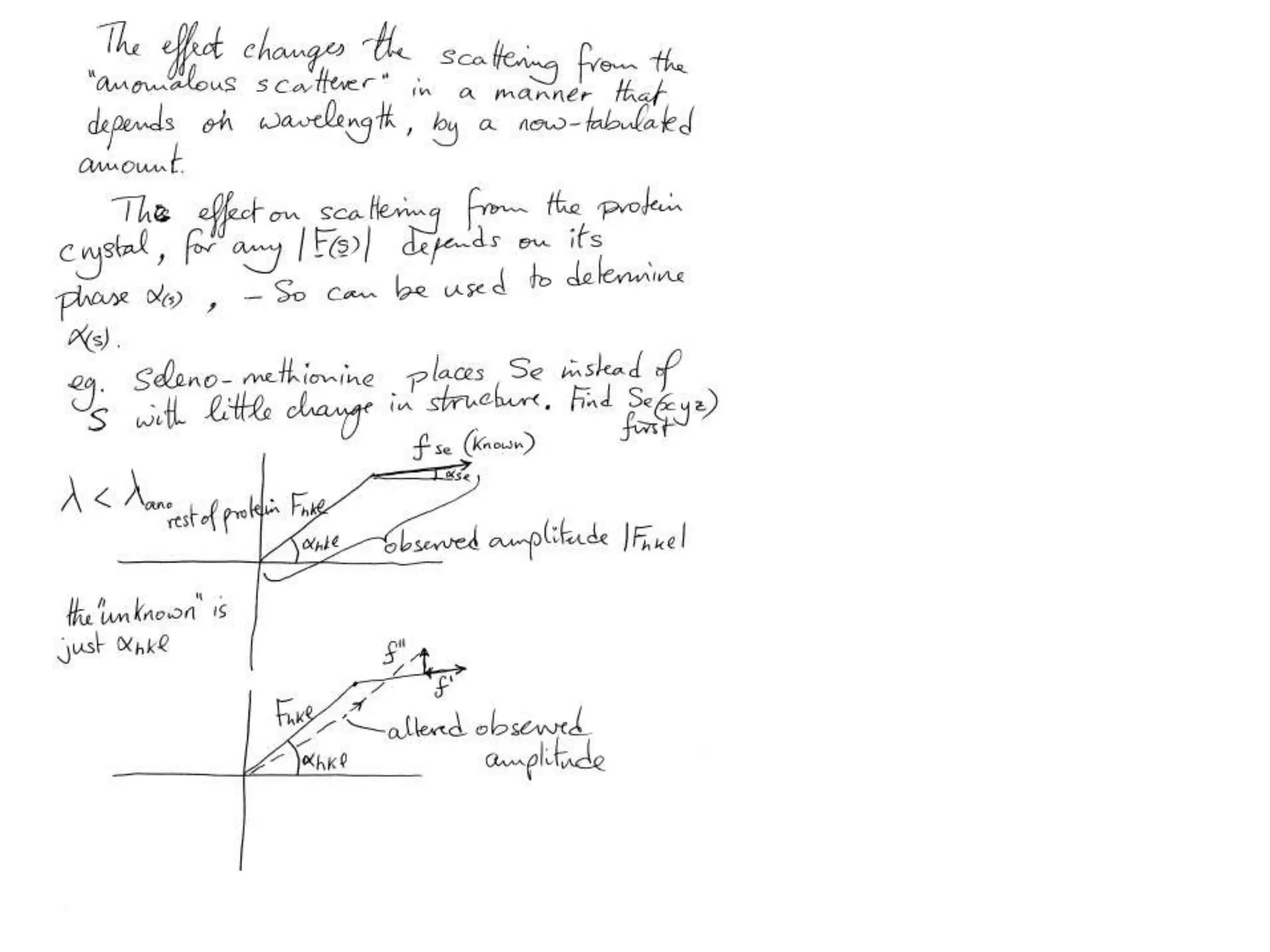
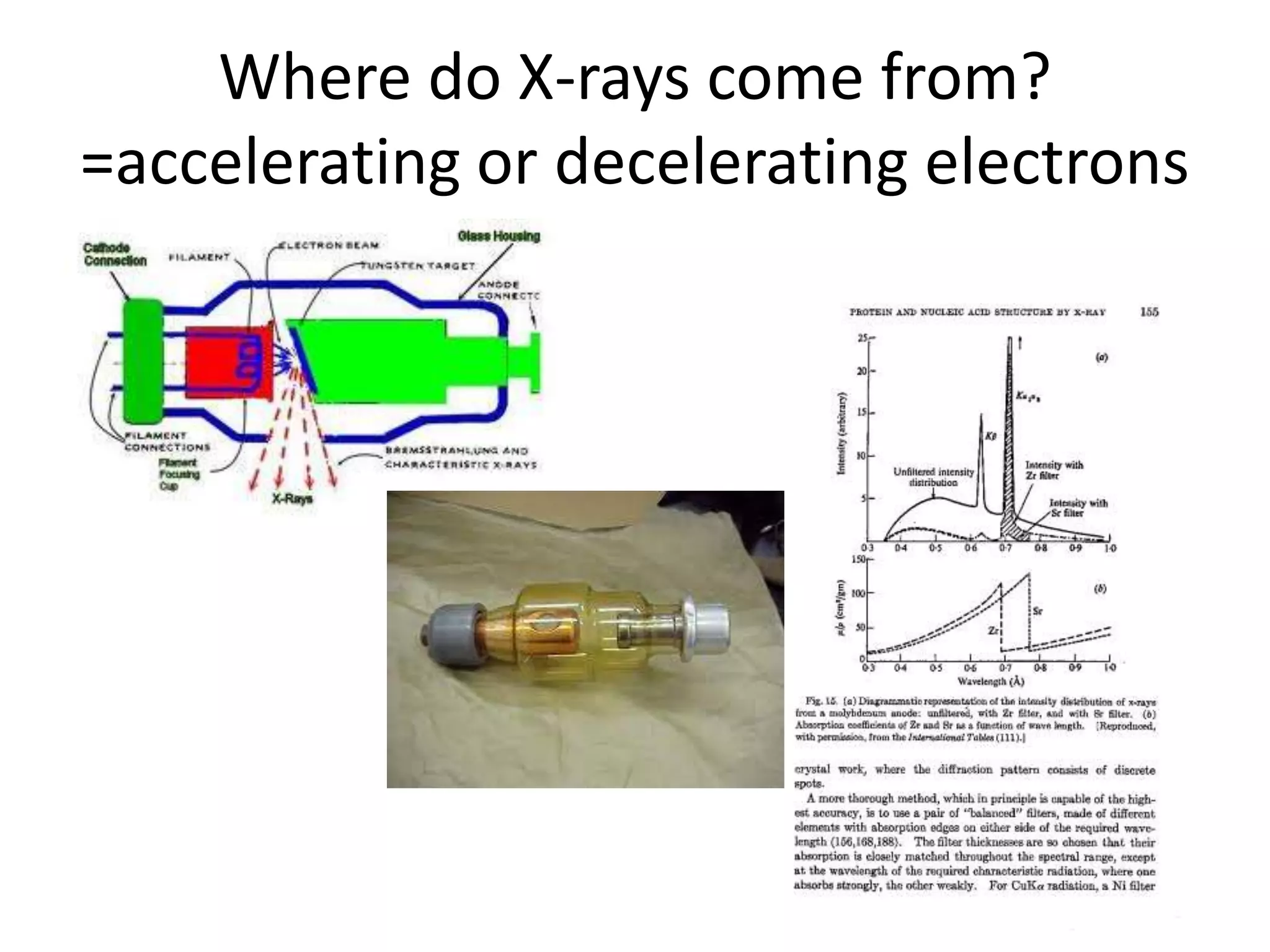
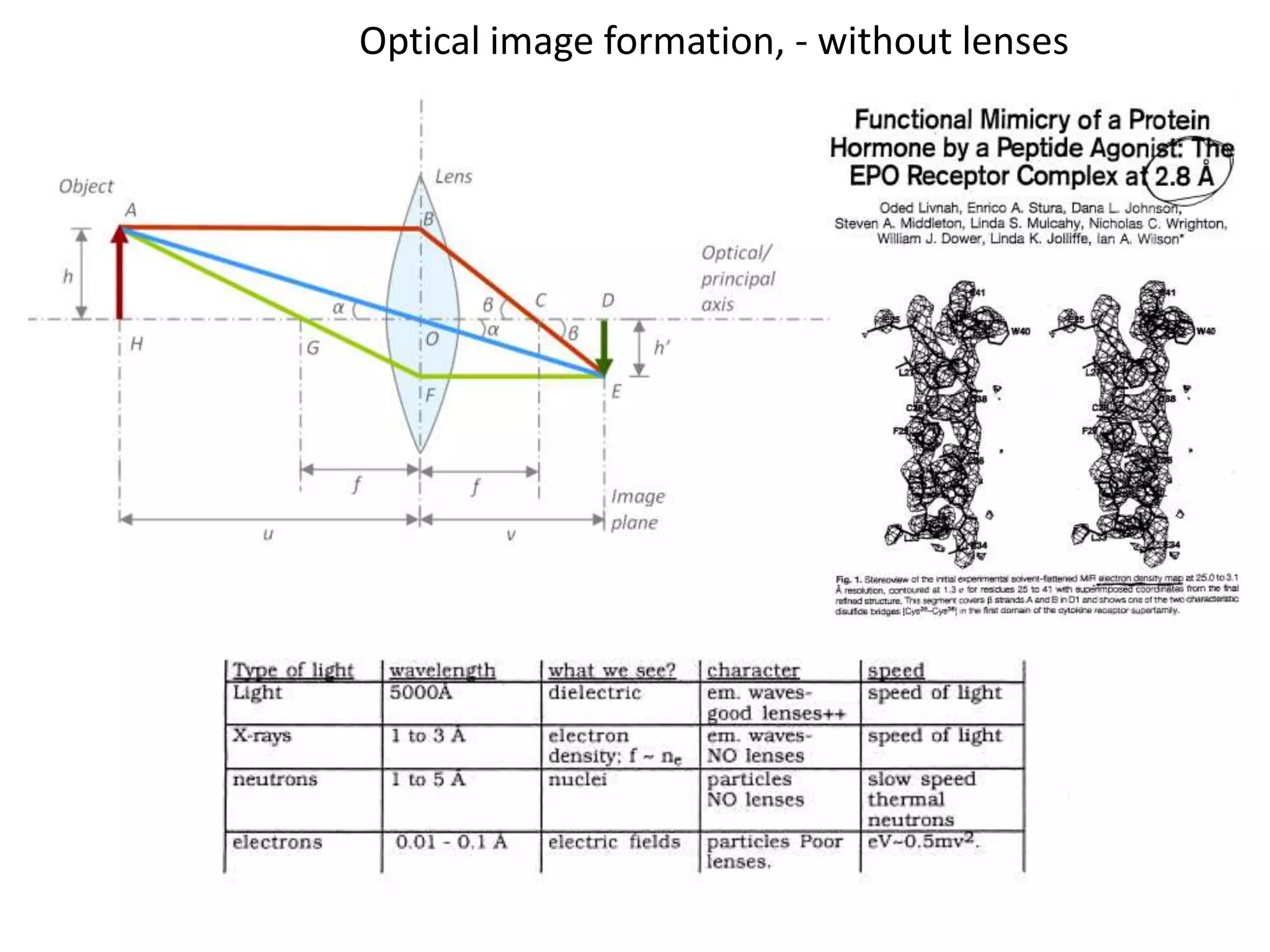
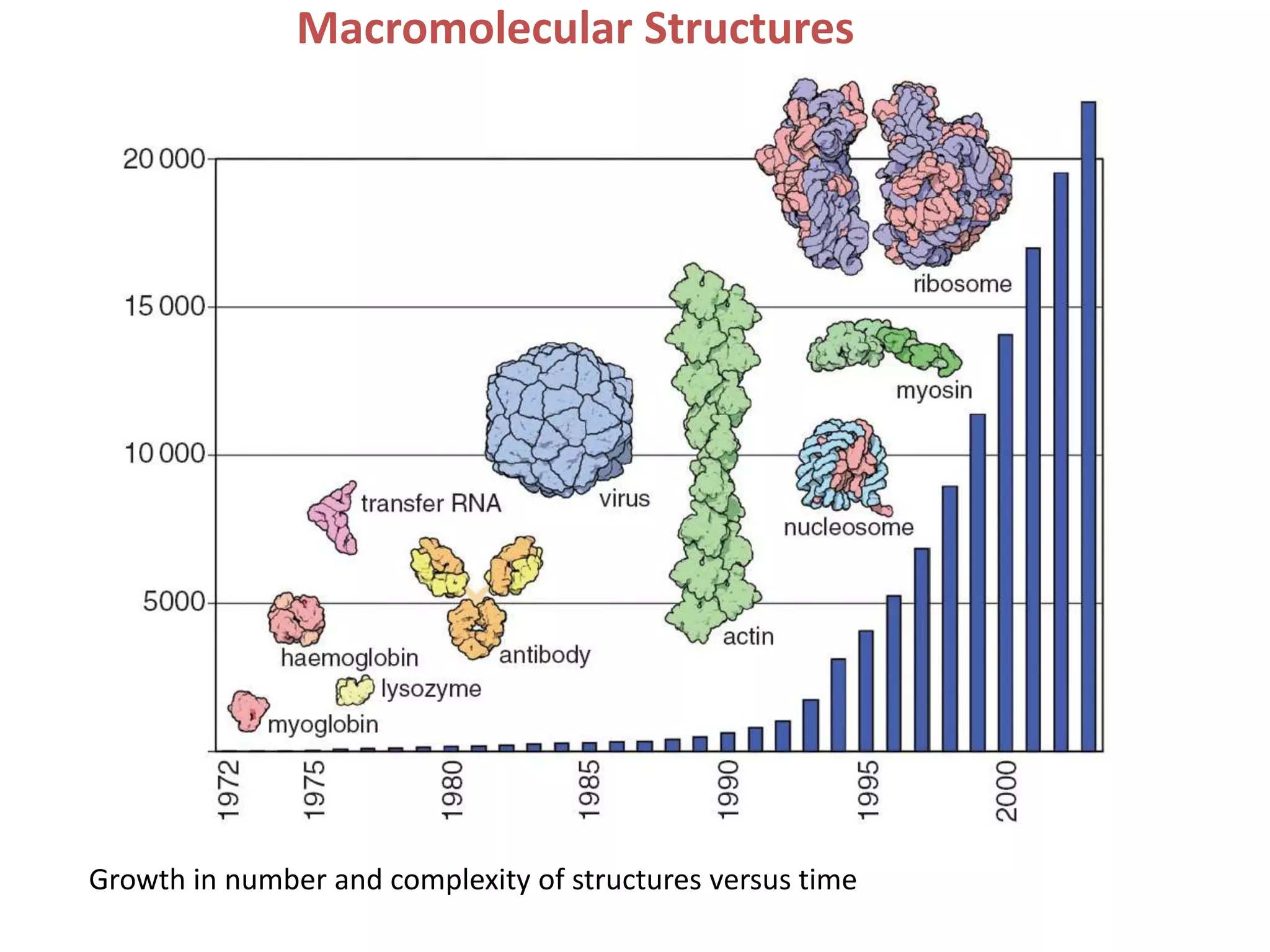
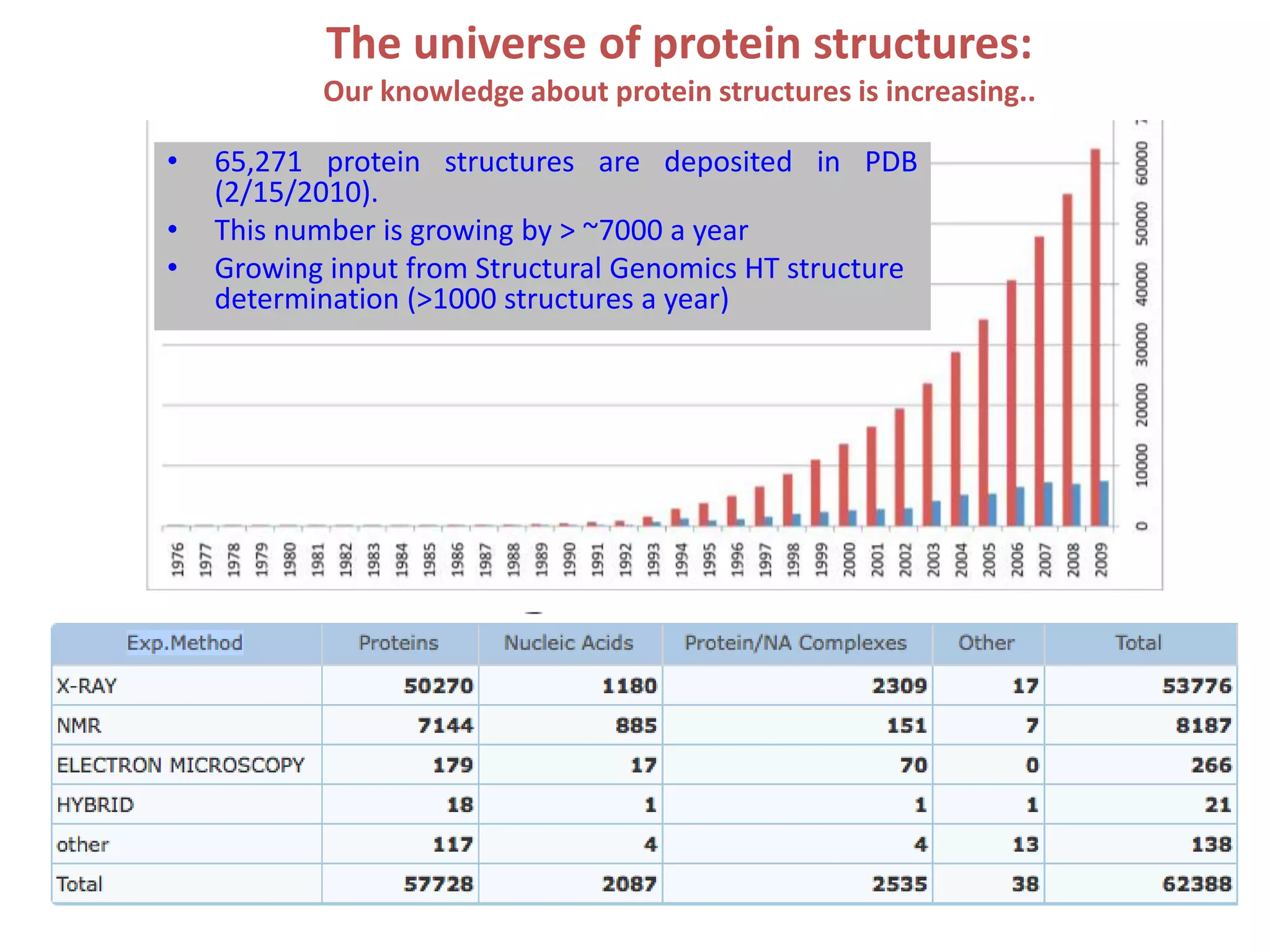
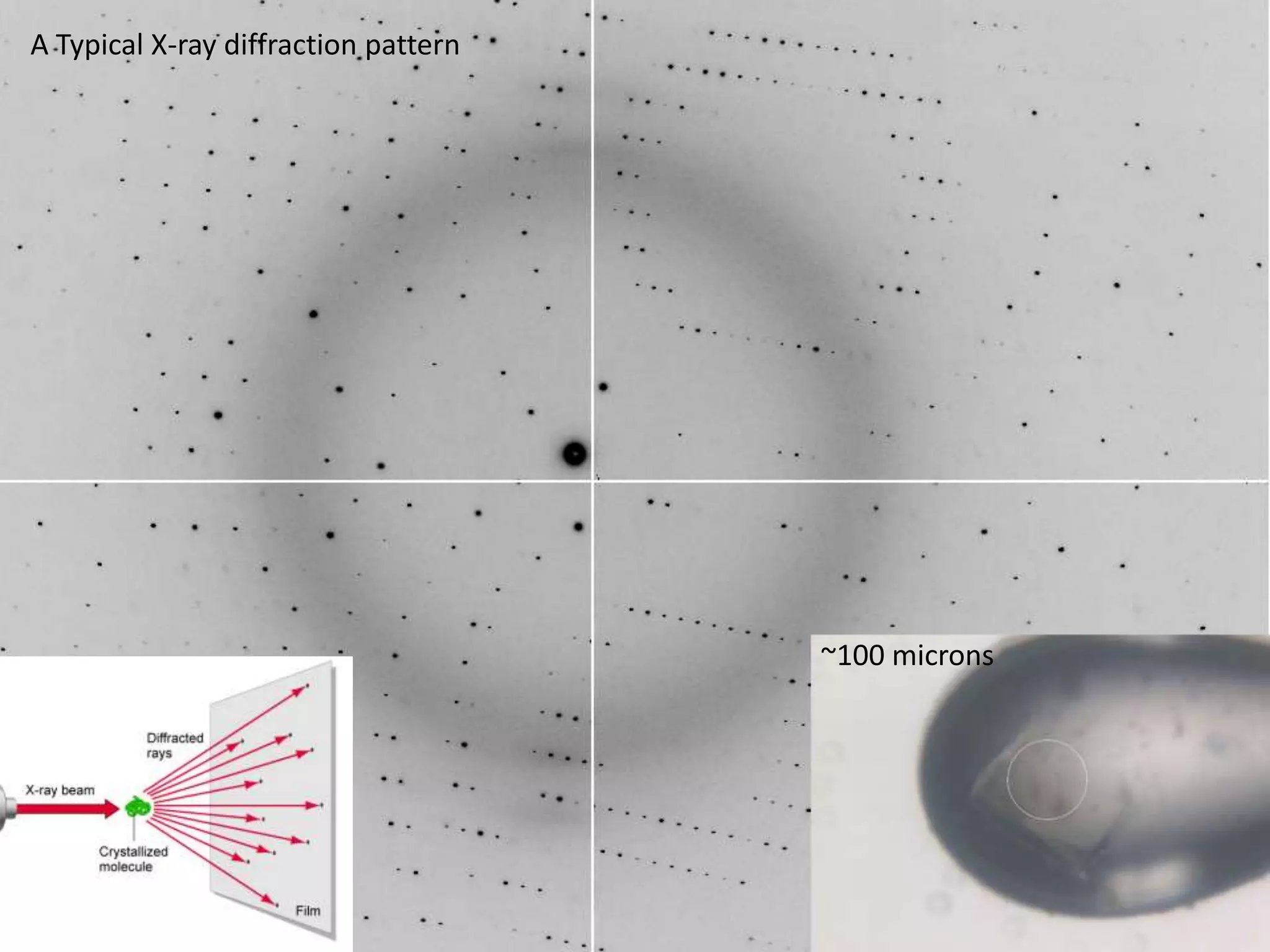
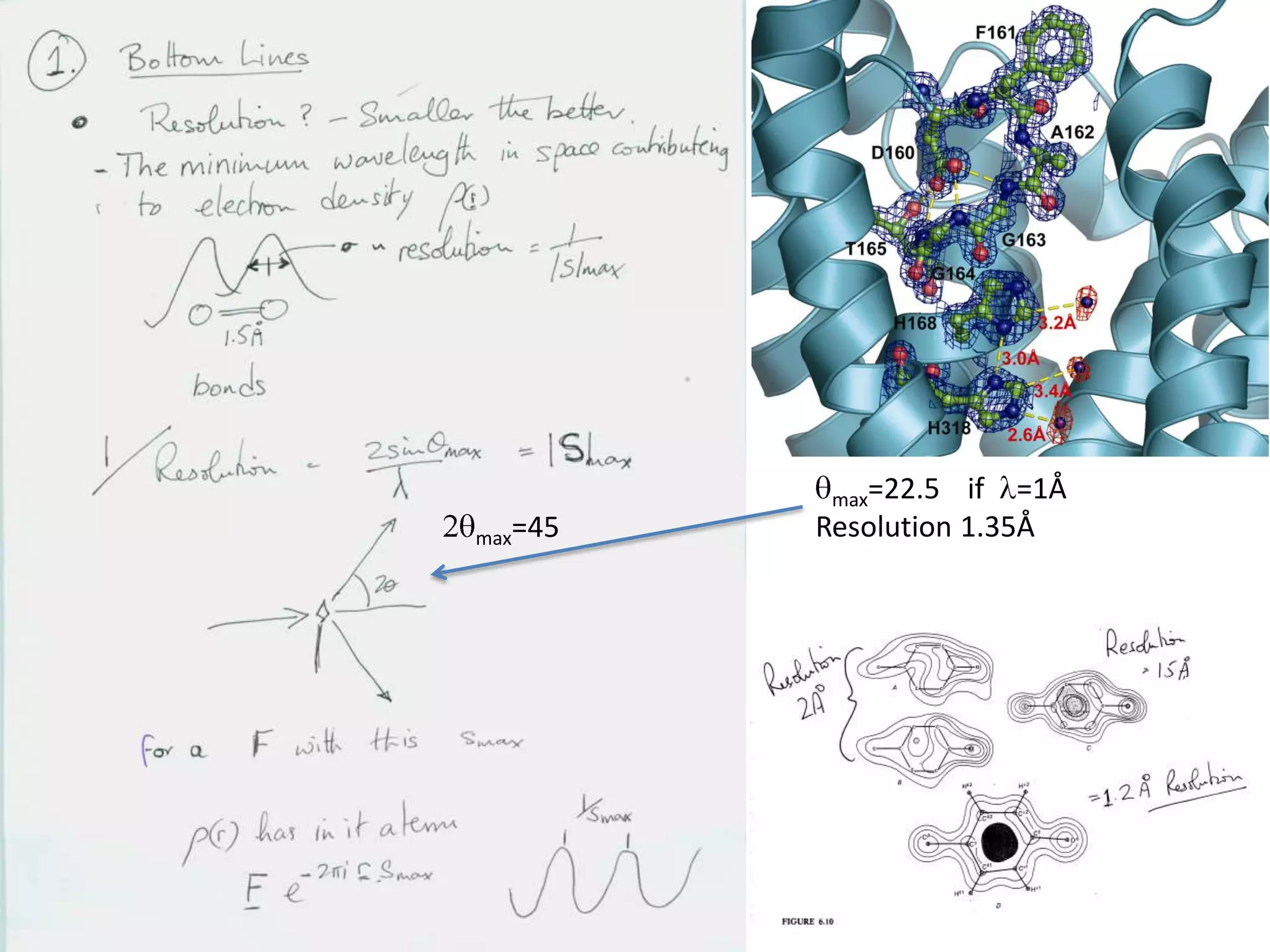
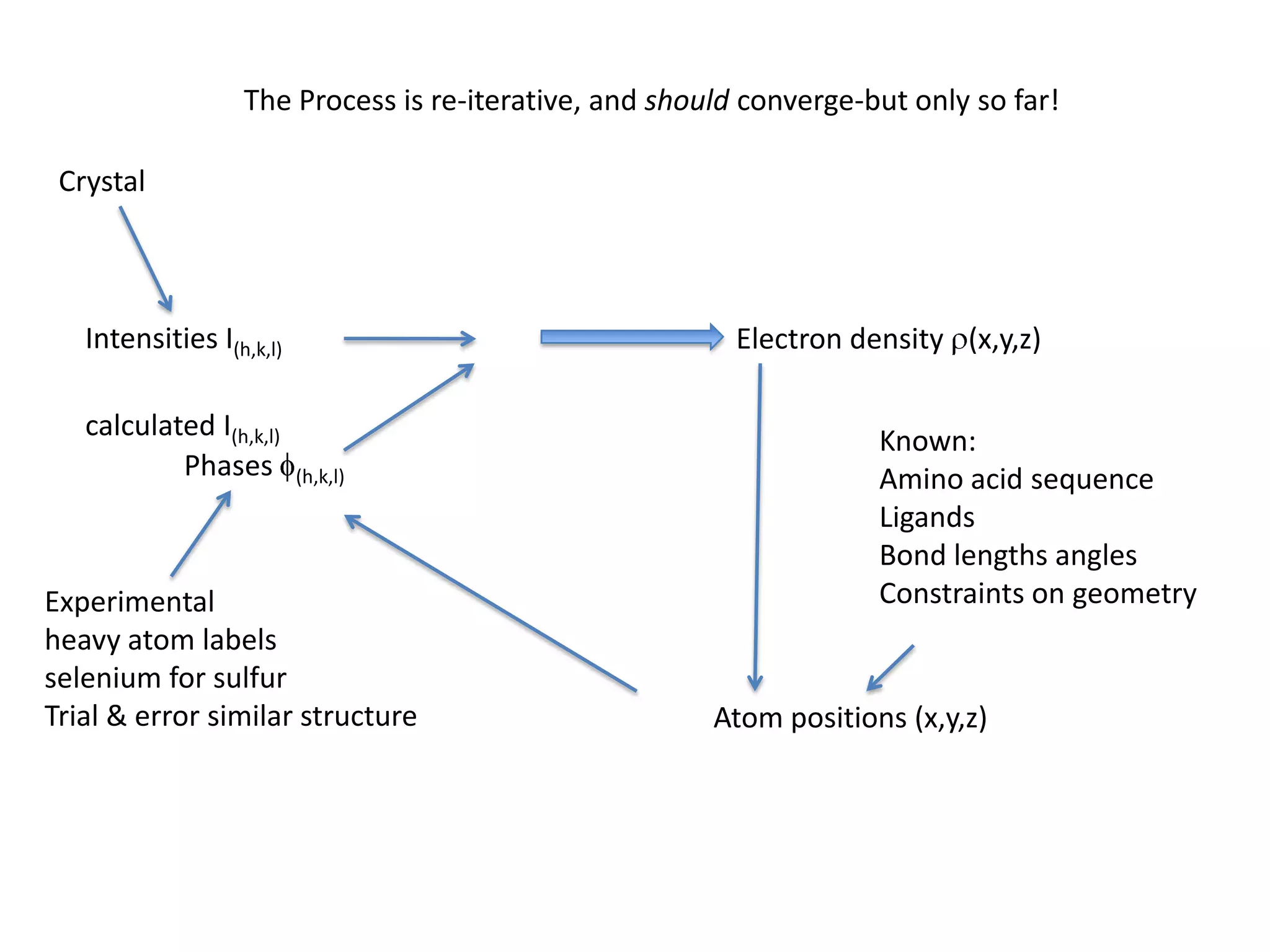
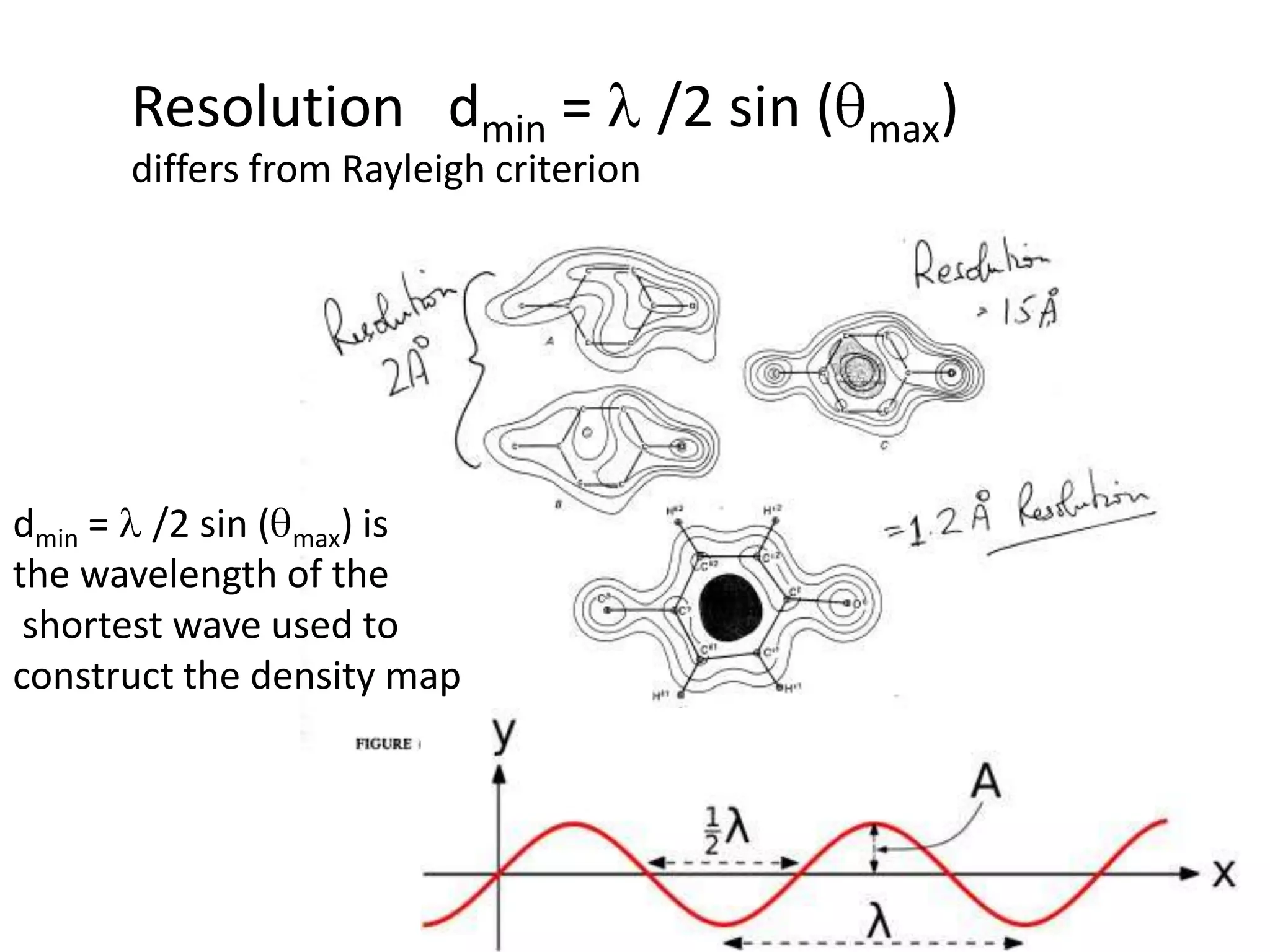
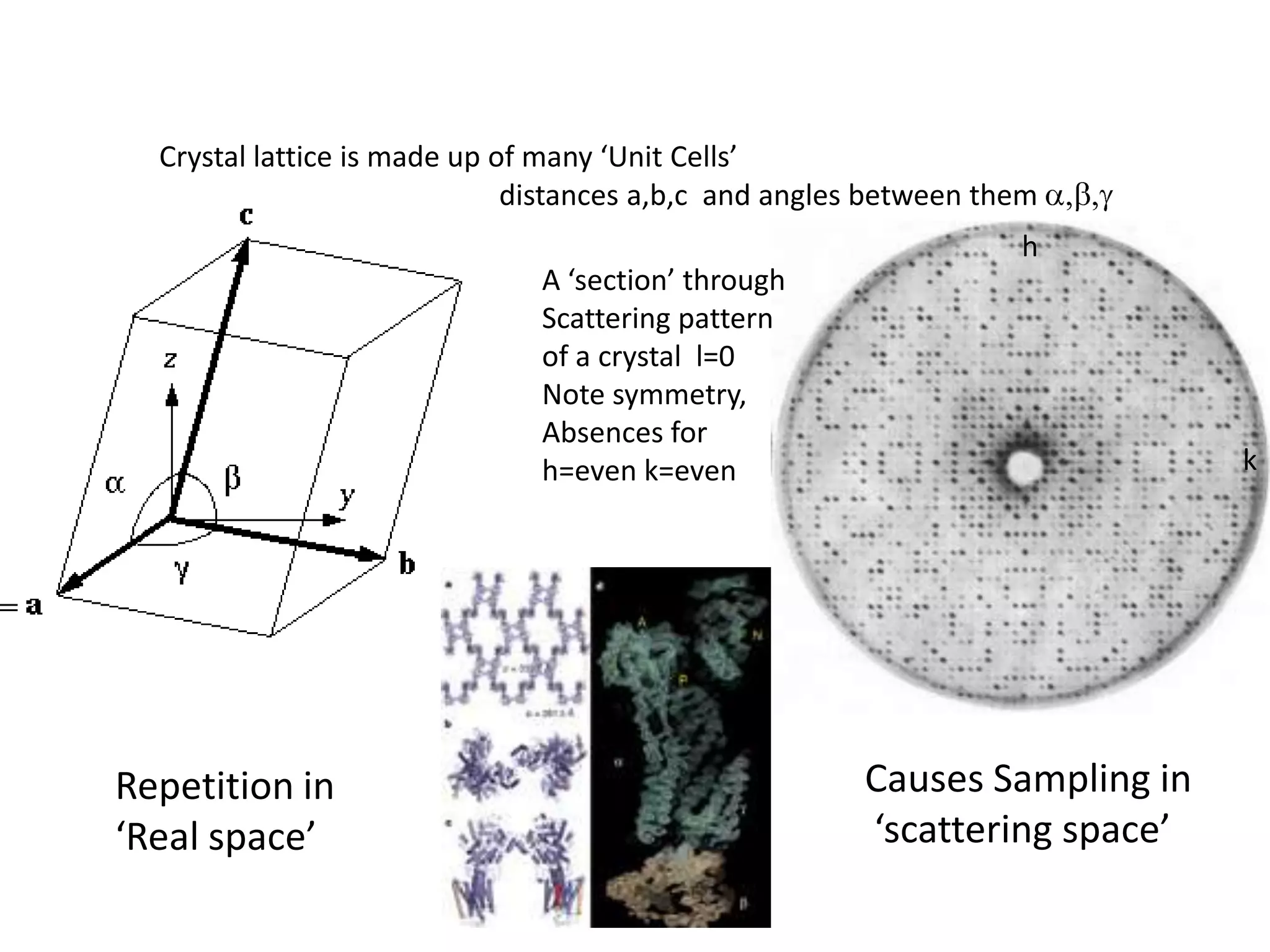
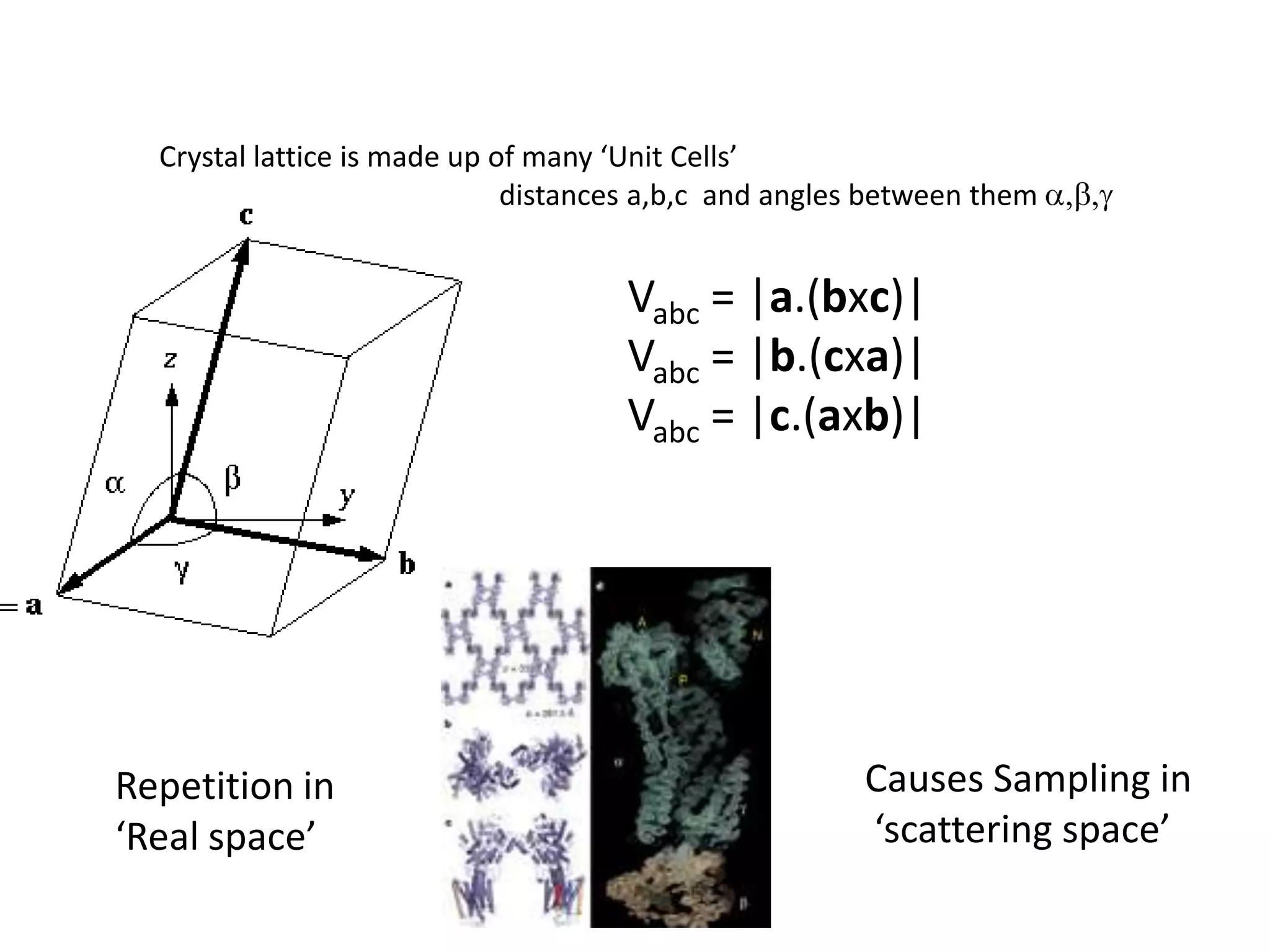
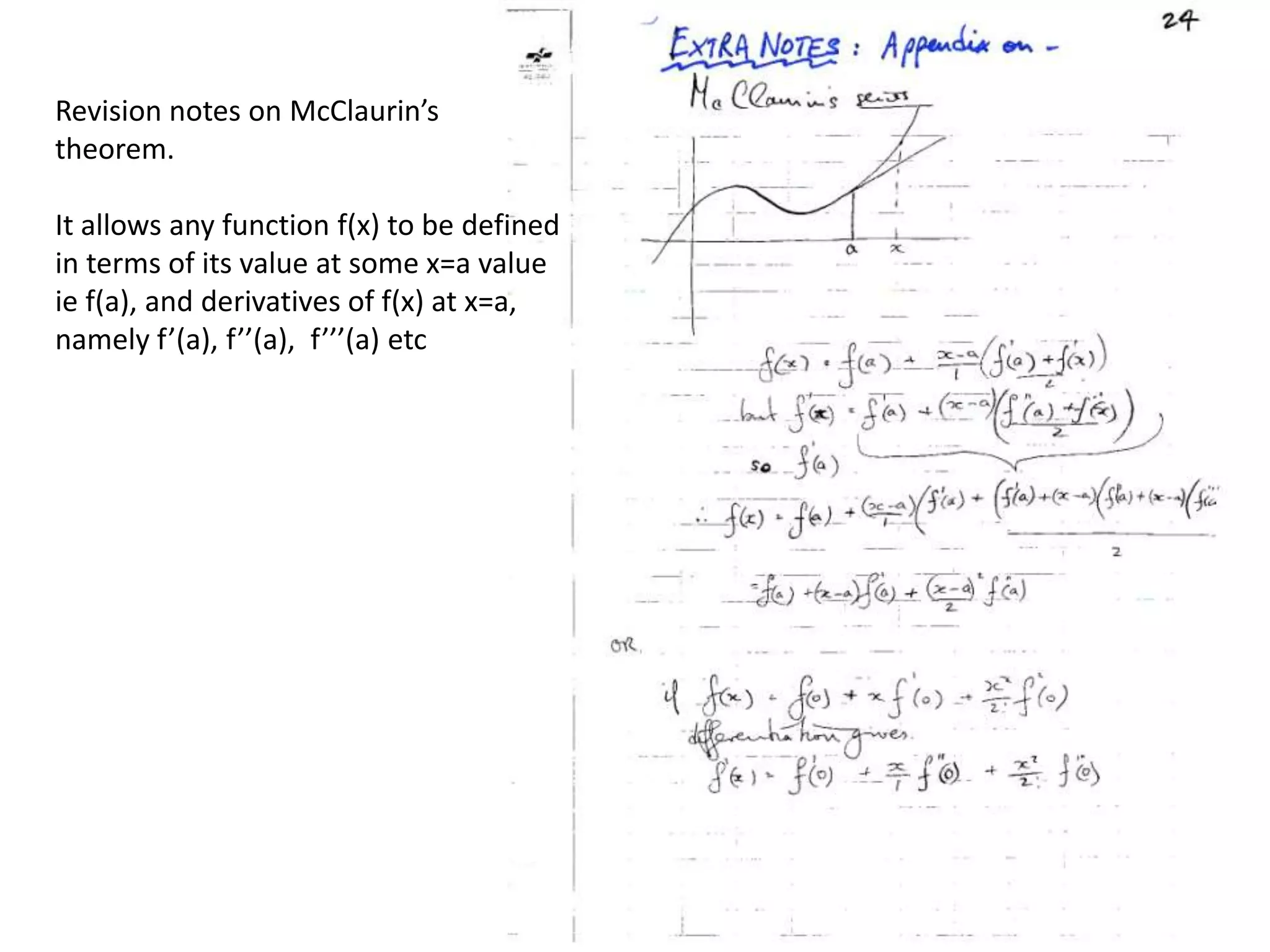
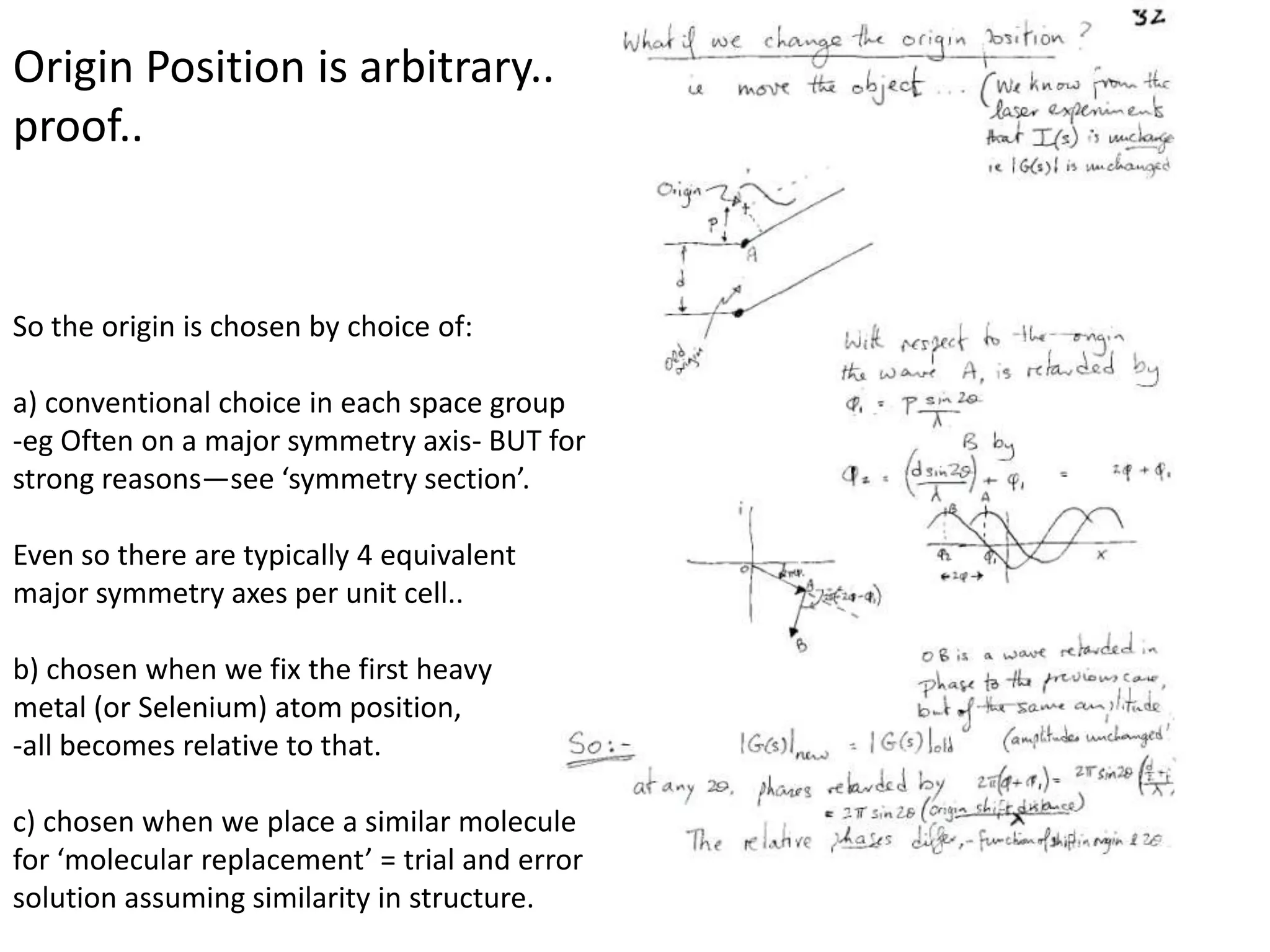
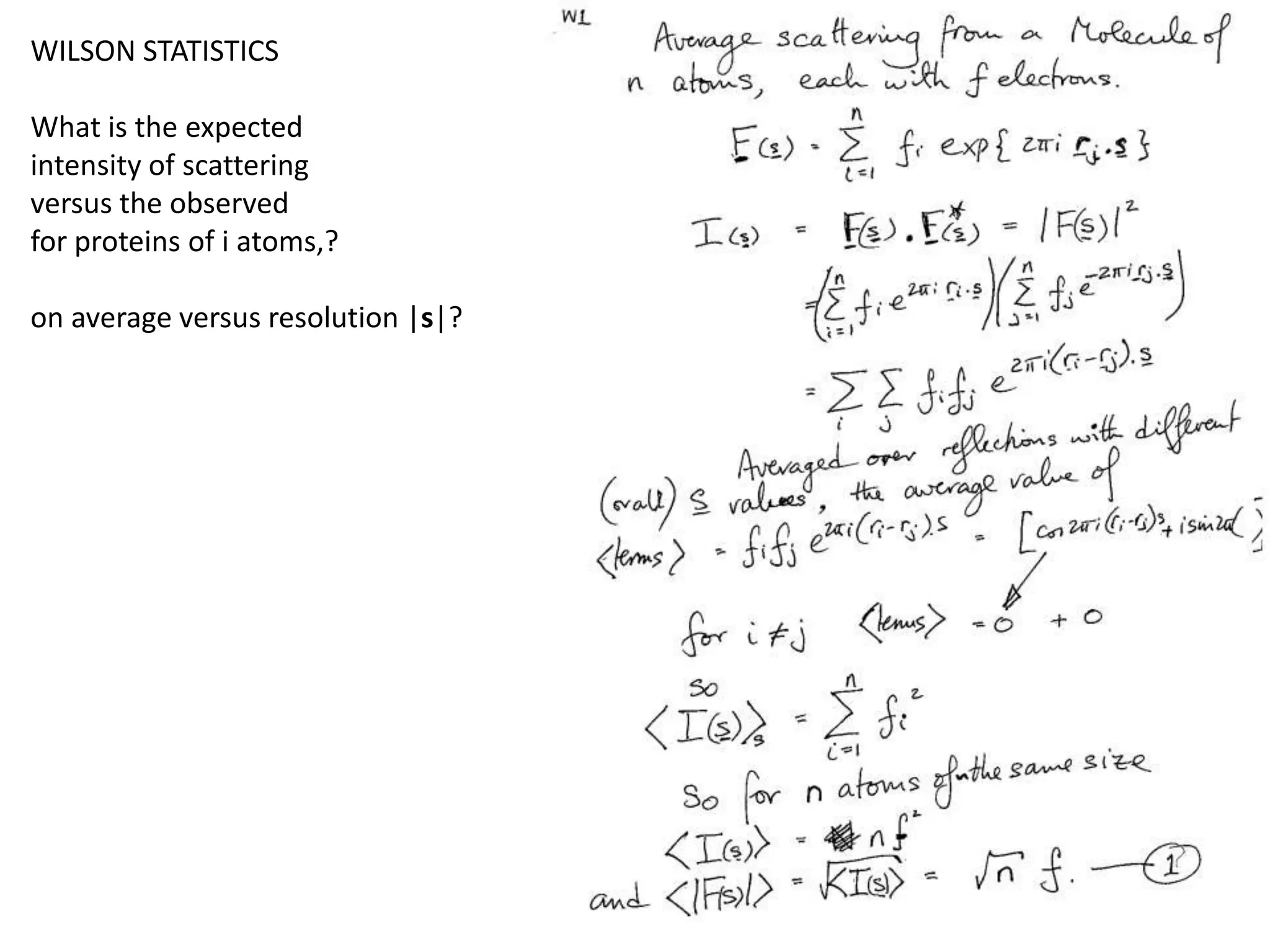
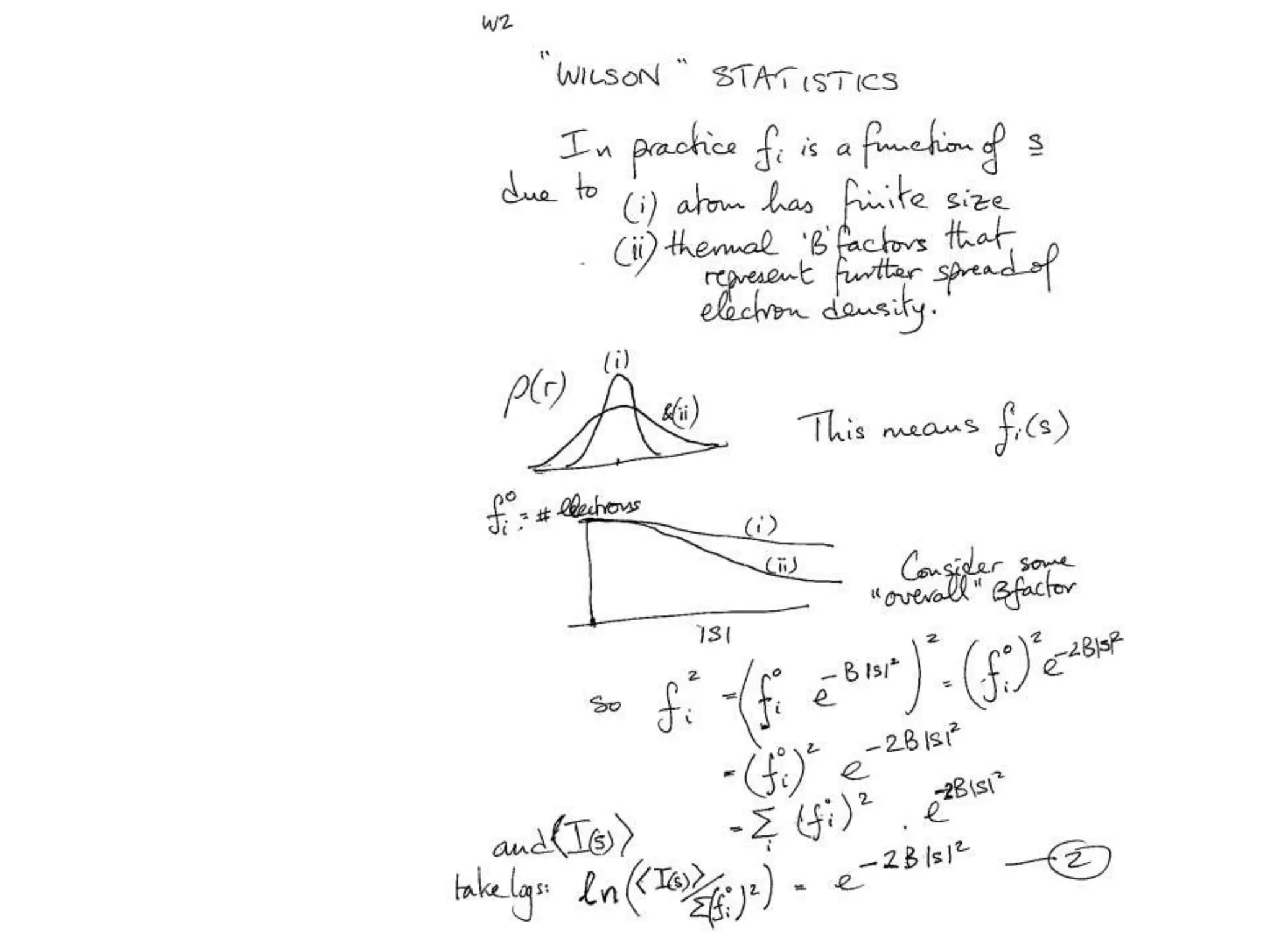
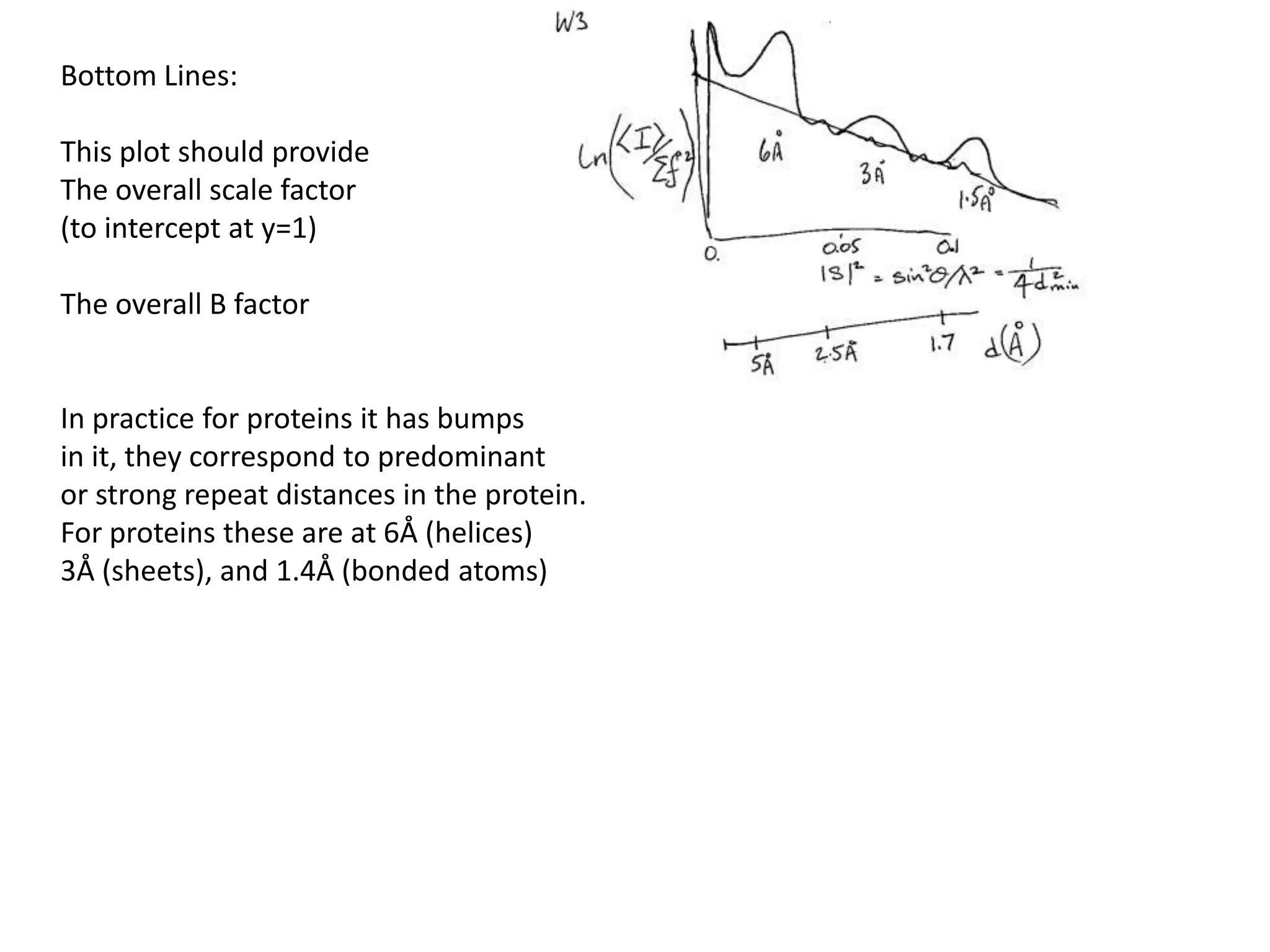
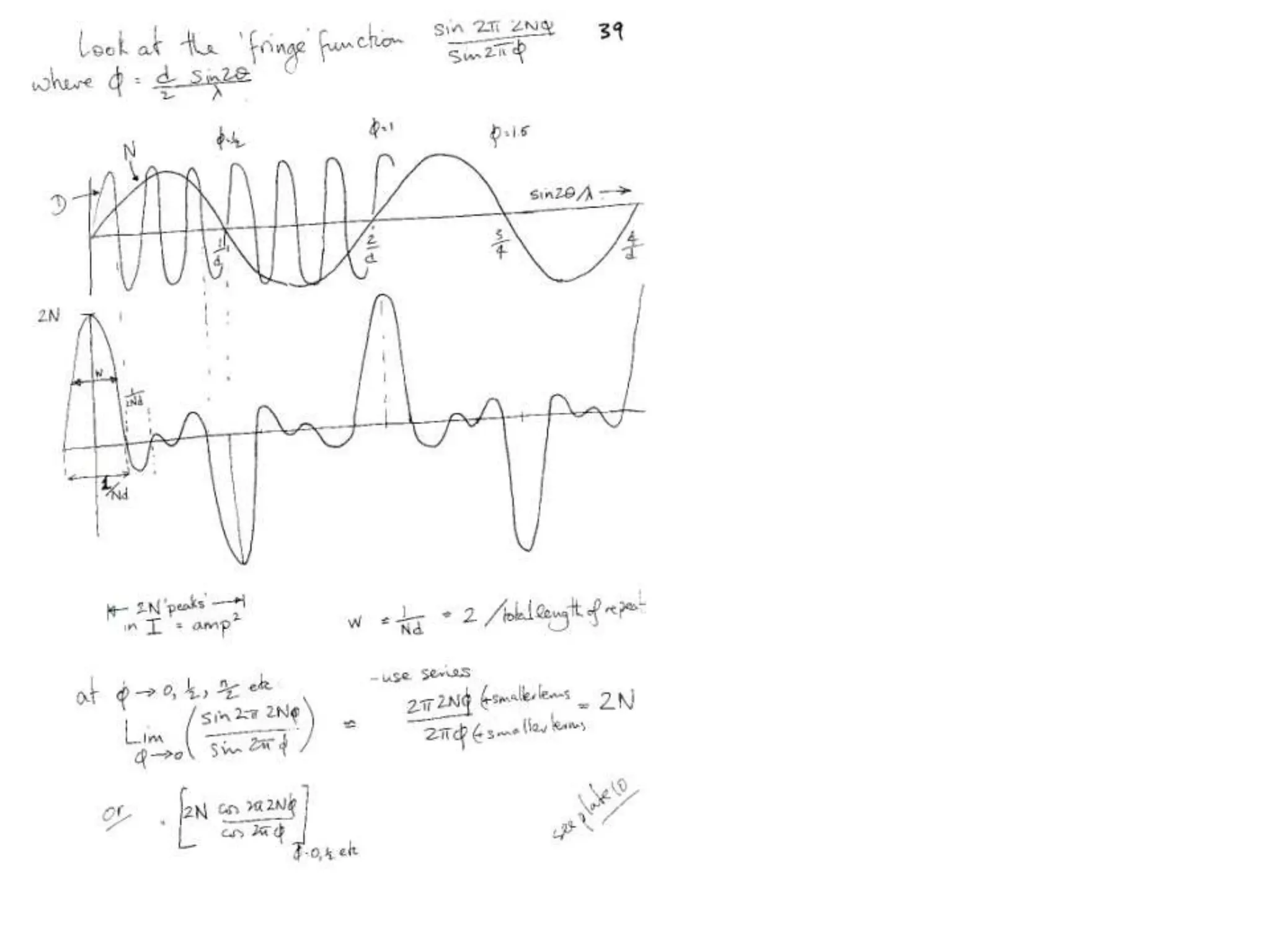
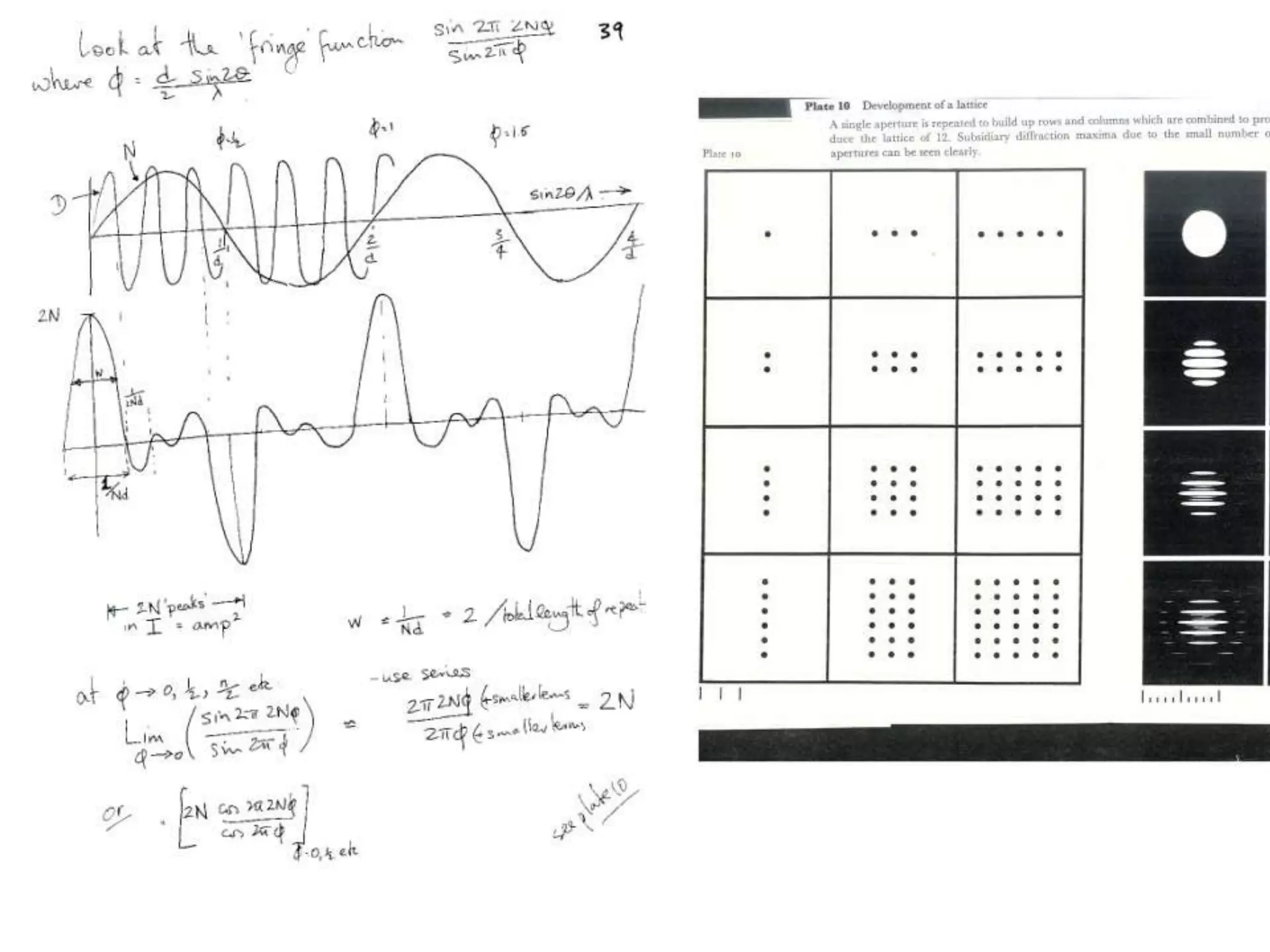
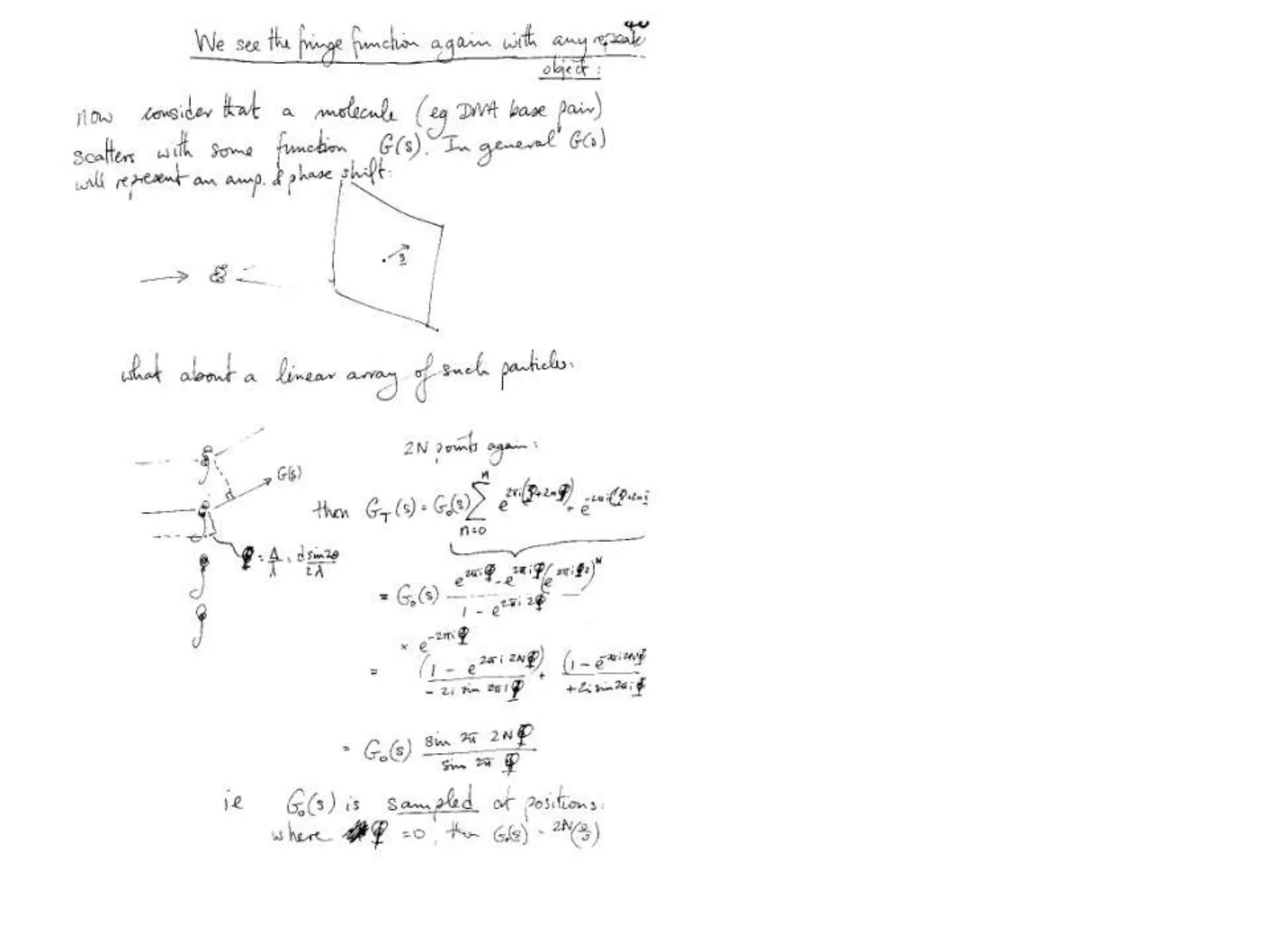
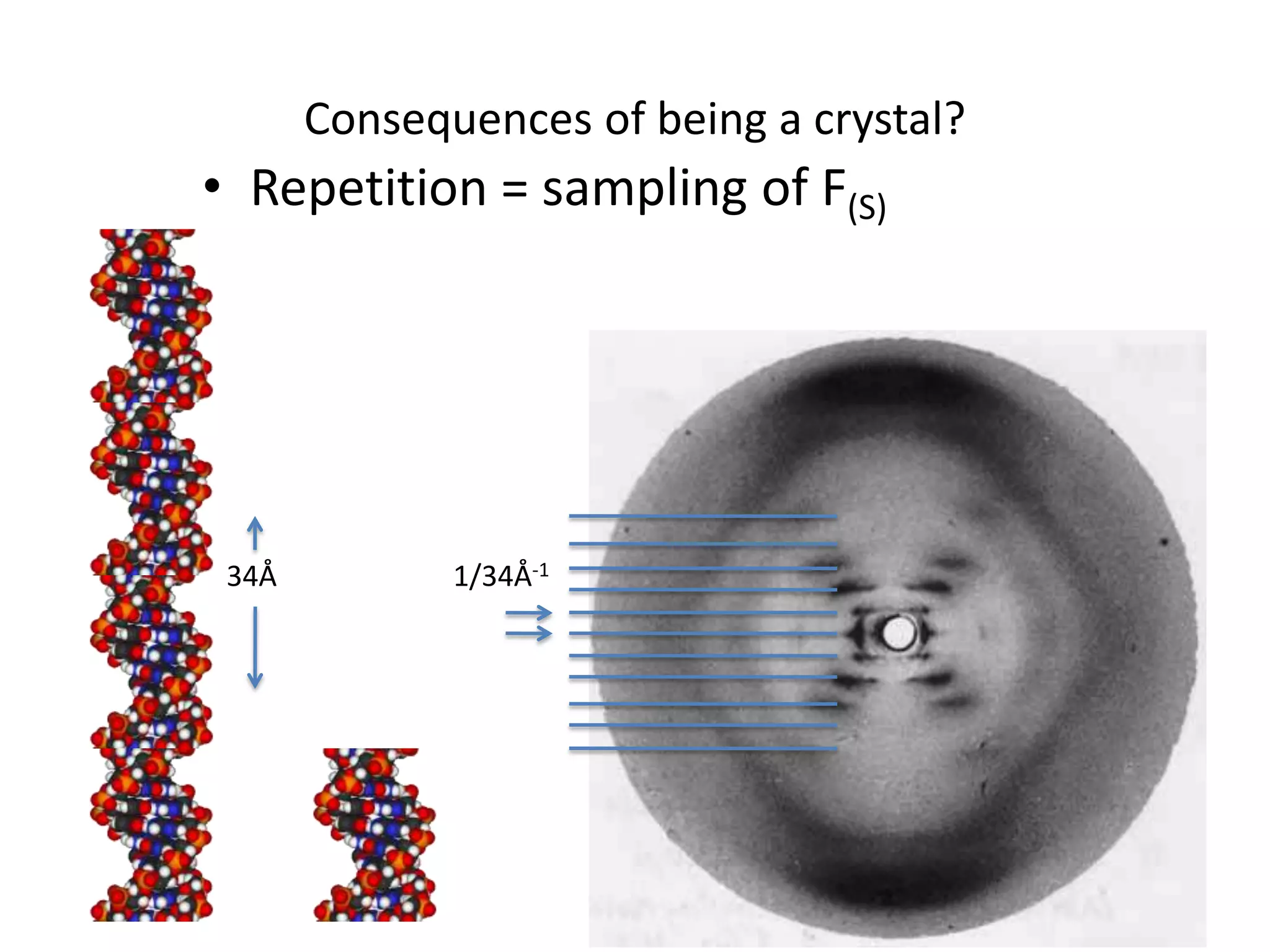
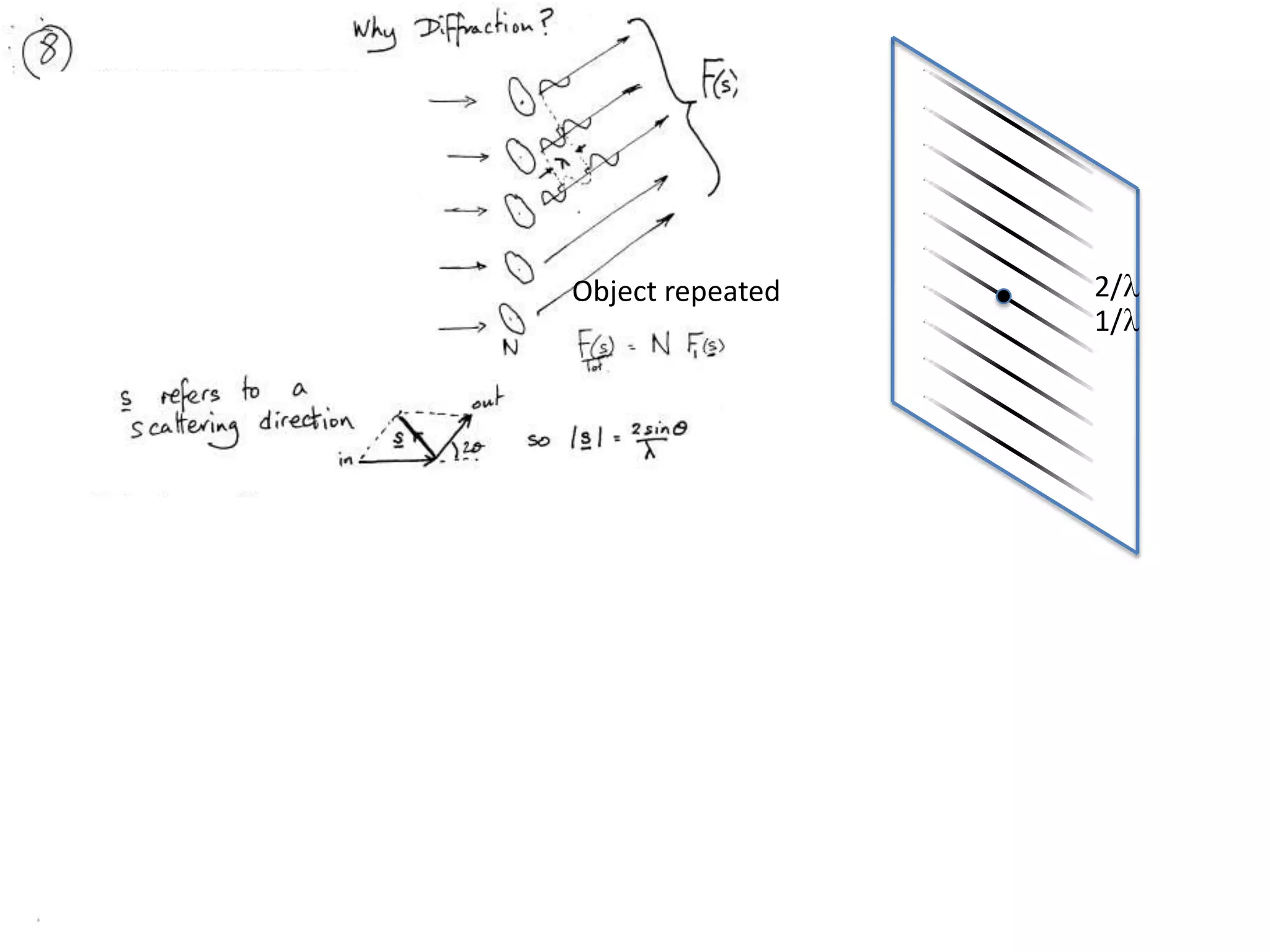
1. The document provides resources and information for determining protein crystal structures using x-ray crystallography. It discusses topics like crystal lattices, diffraction, the phase problem, and structure refinement.
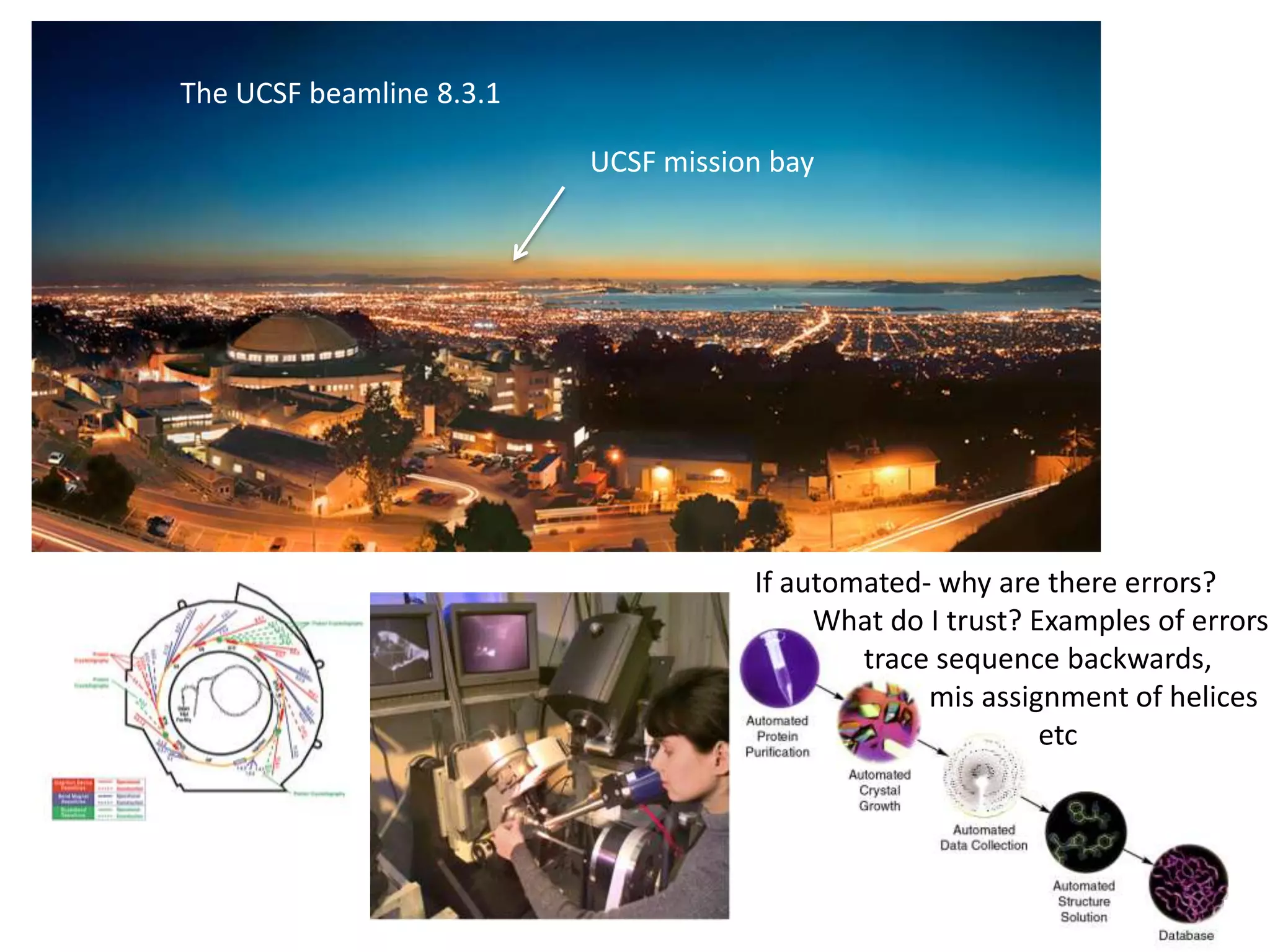
2. Key resources mentioned include the Advanced Light Source for collecting diffraction data, and computing software for analyzing structures.
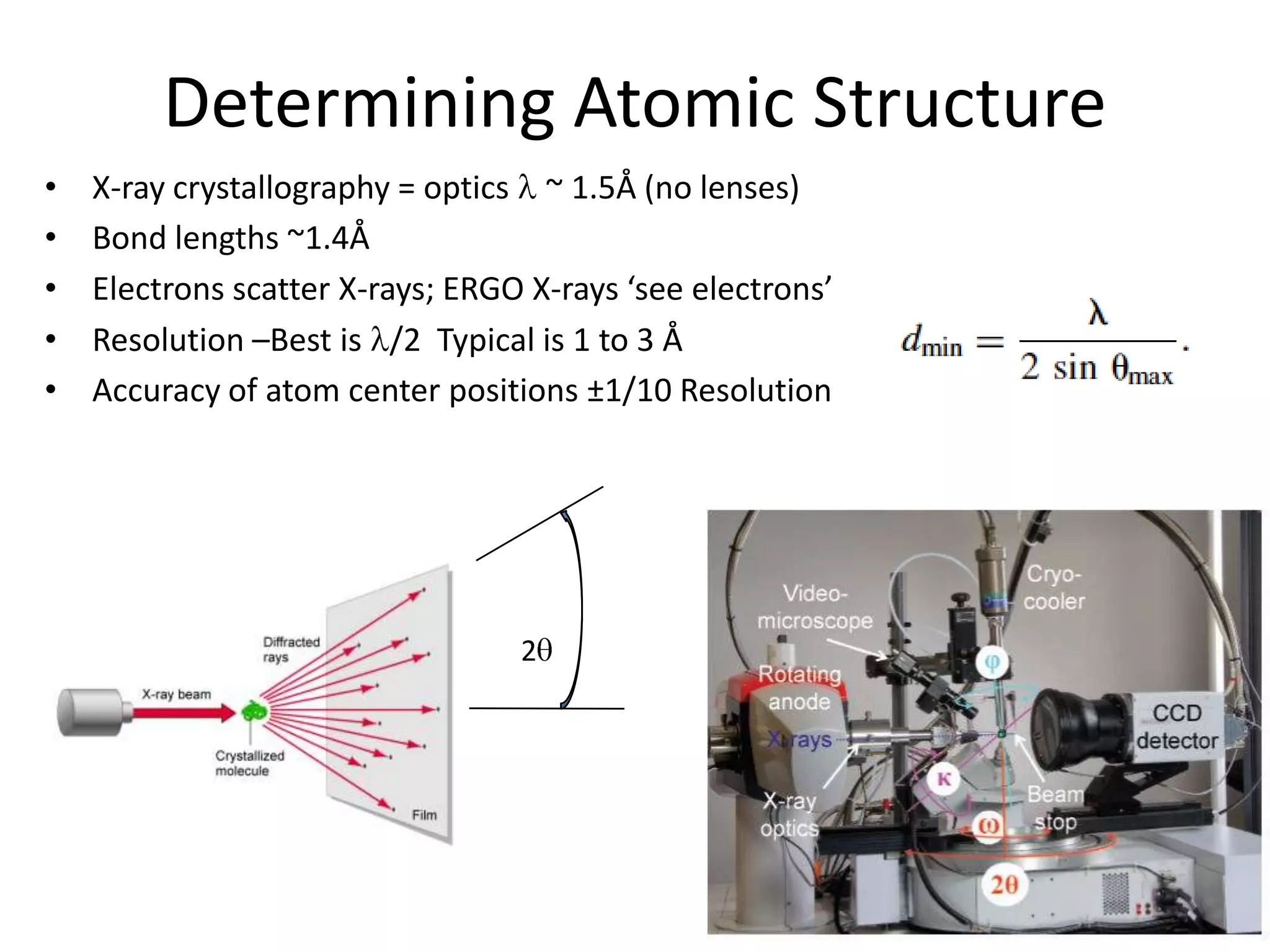
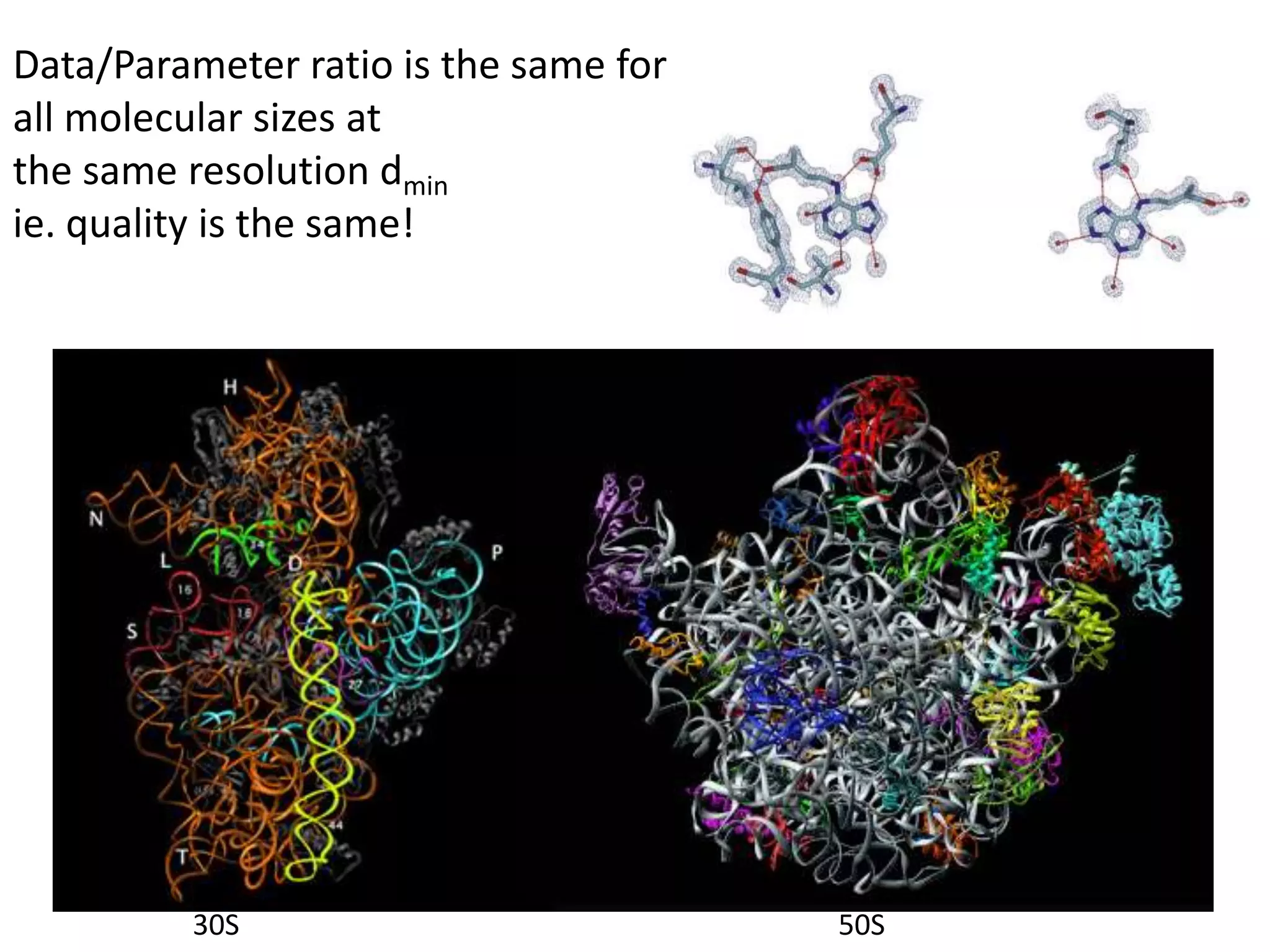
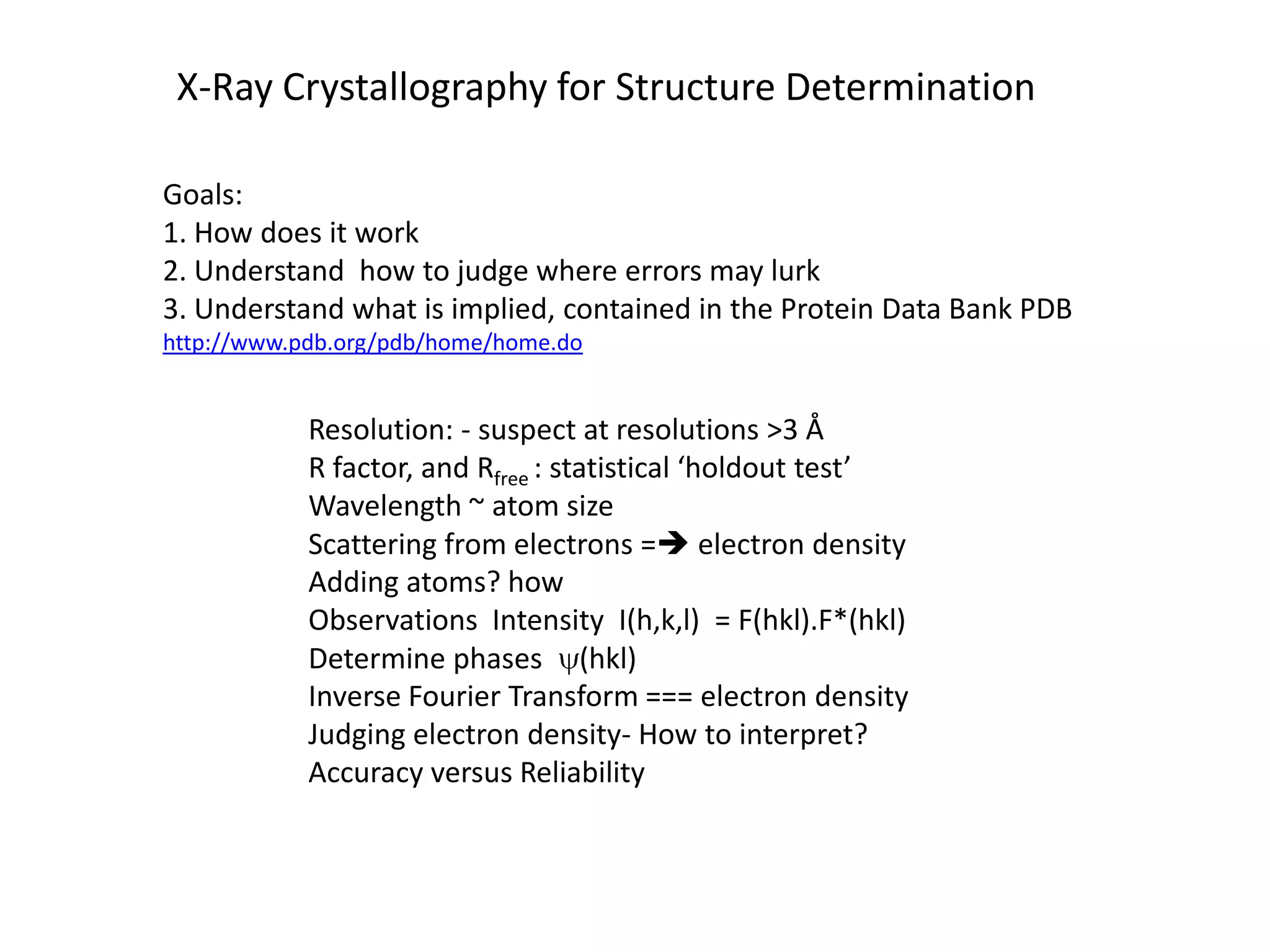
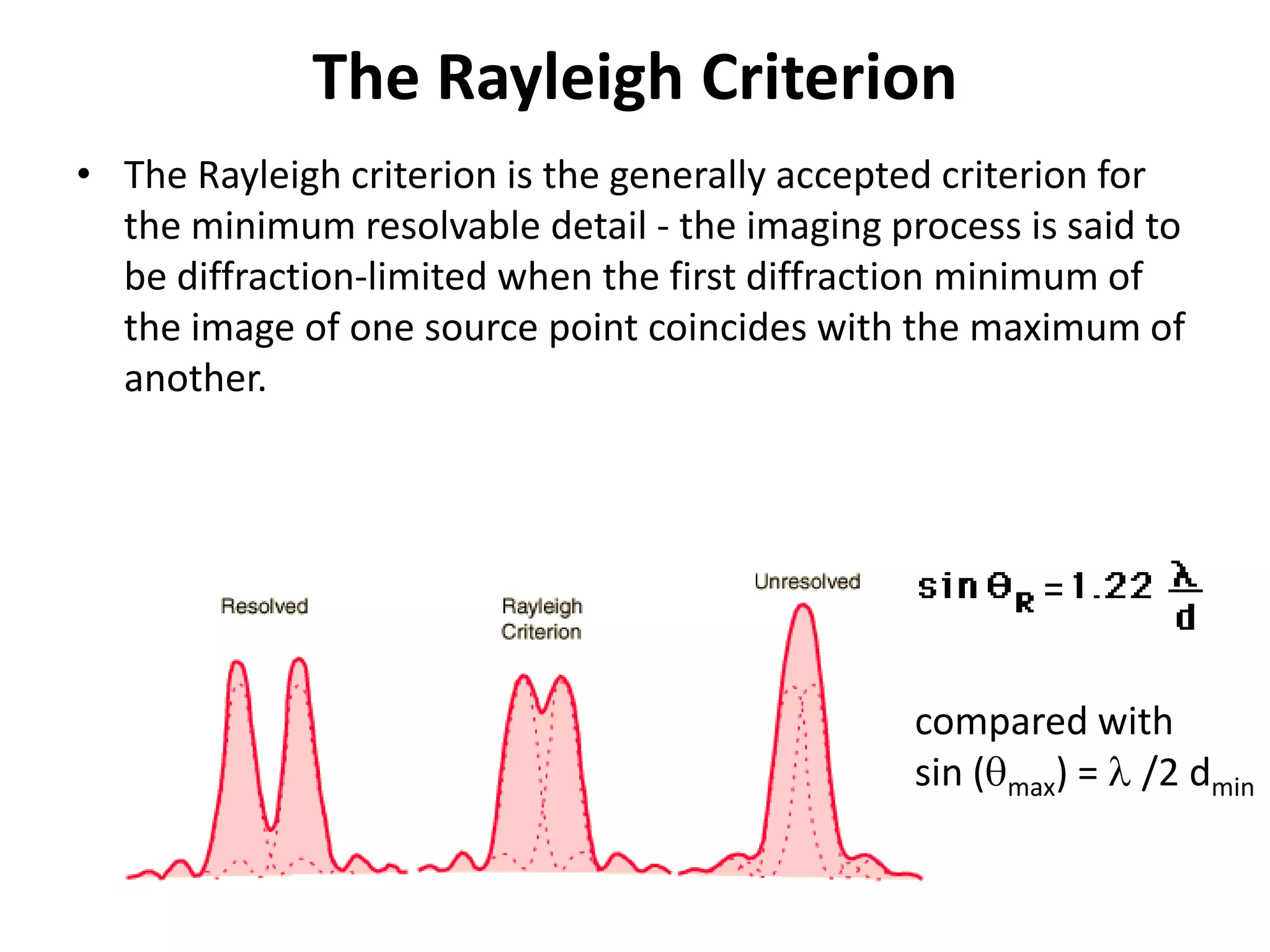
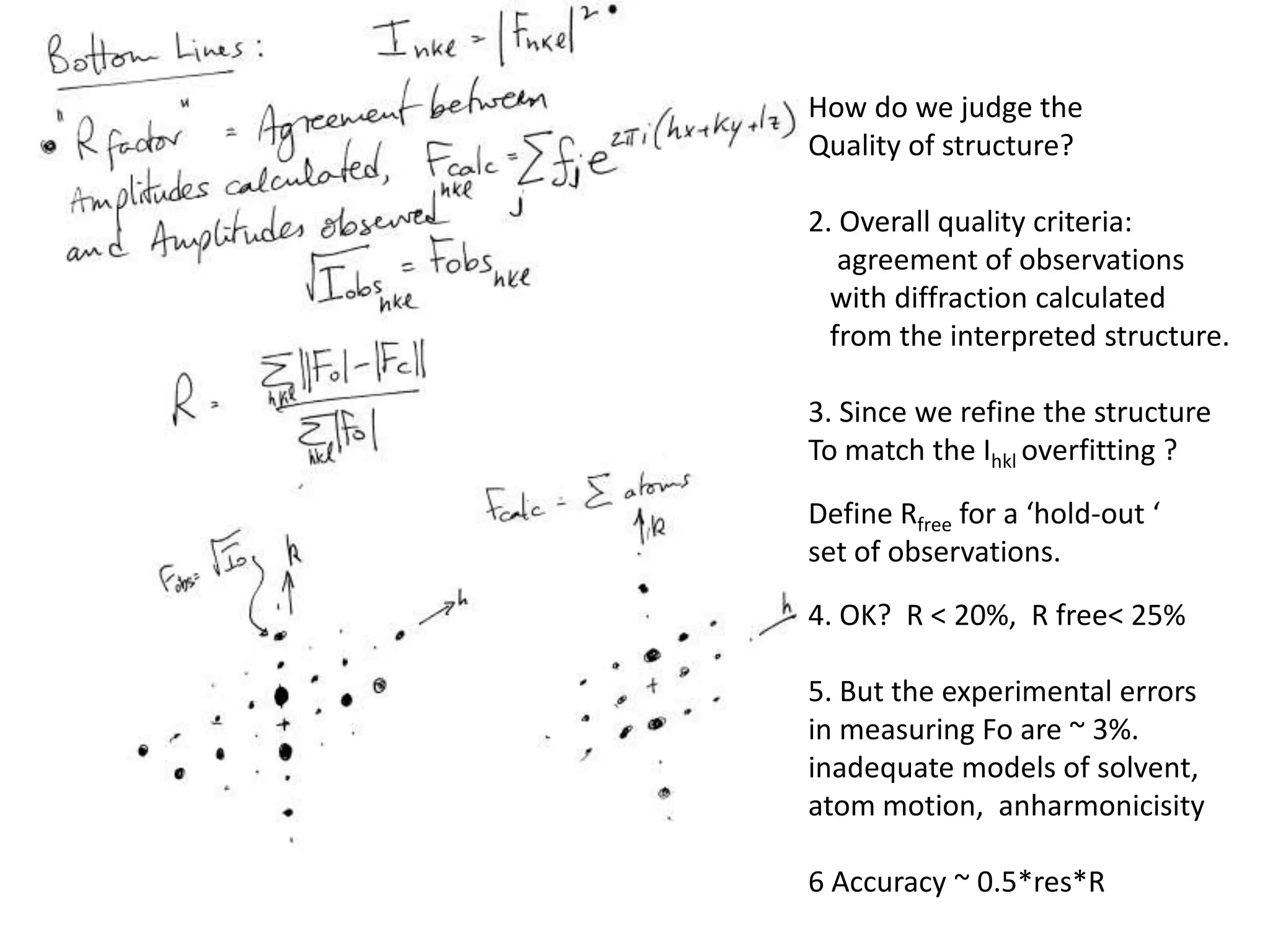
3. The goals of x-ray crystallography are outlined as determining how the technique works, understanding potential sources of error, and what information is contained in the Protein Data Bank structure database.



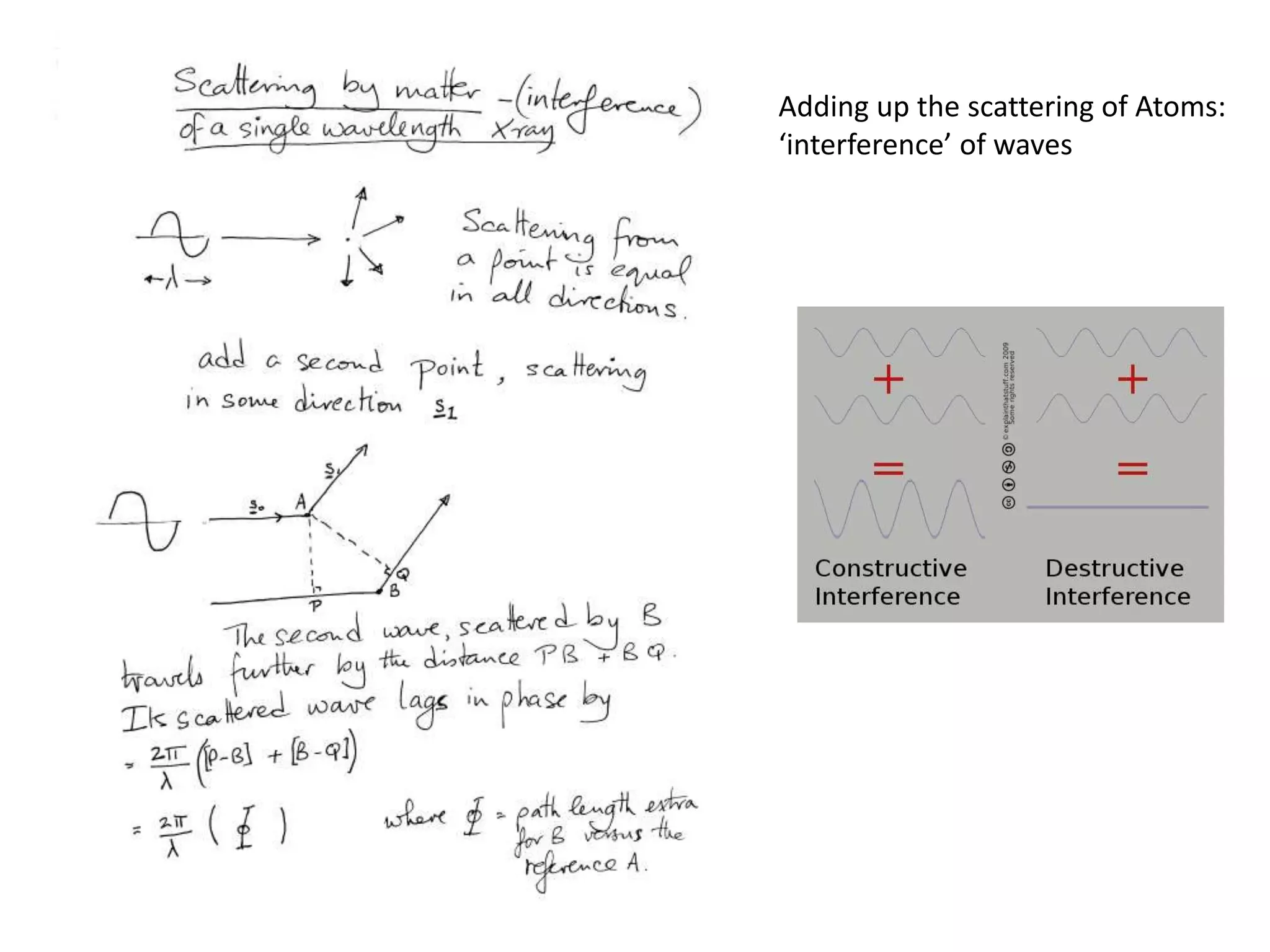
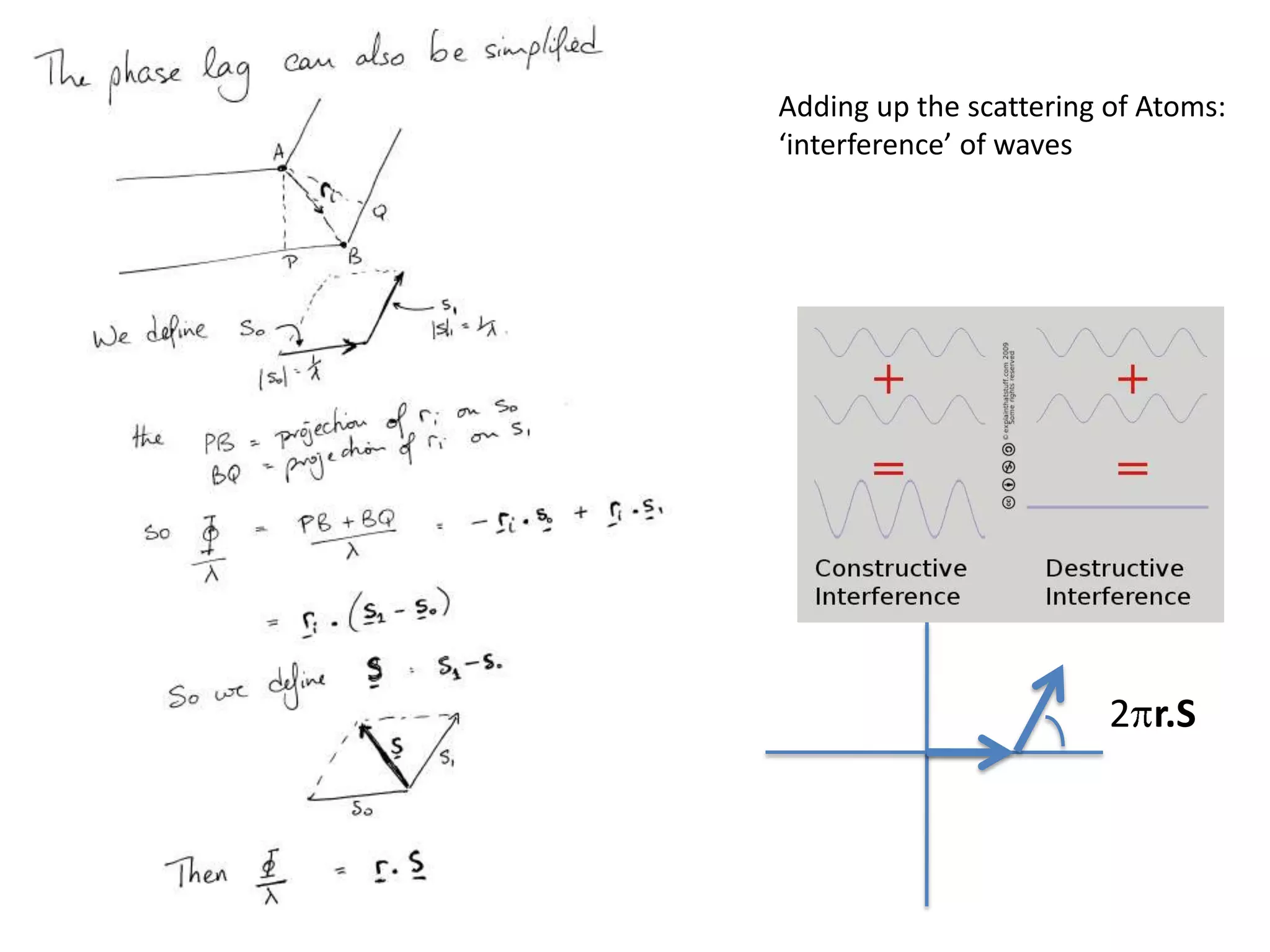
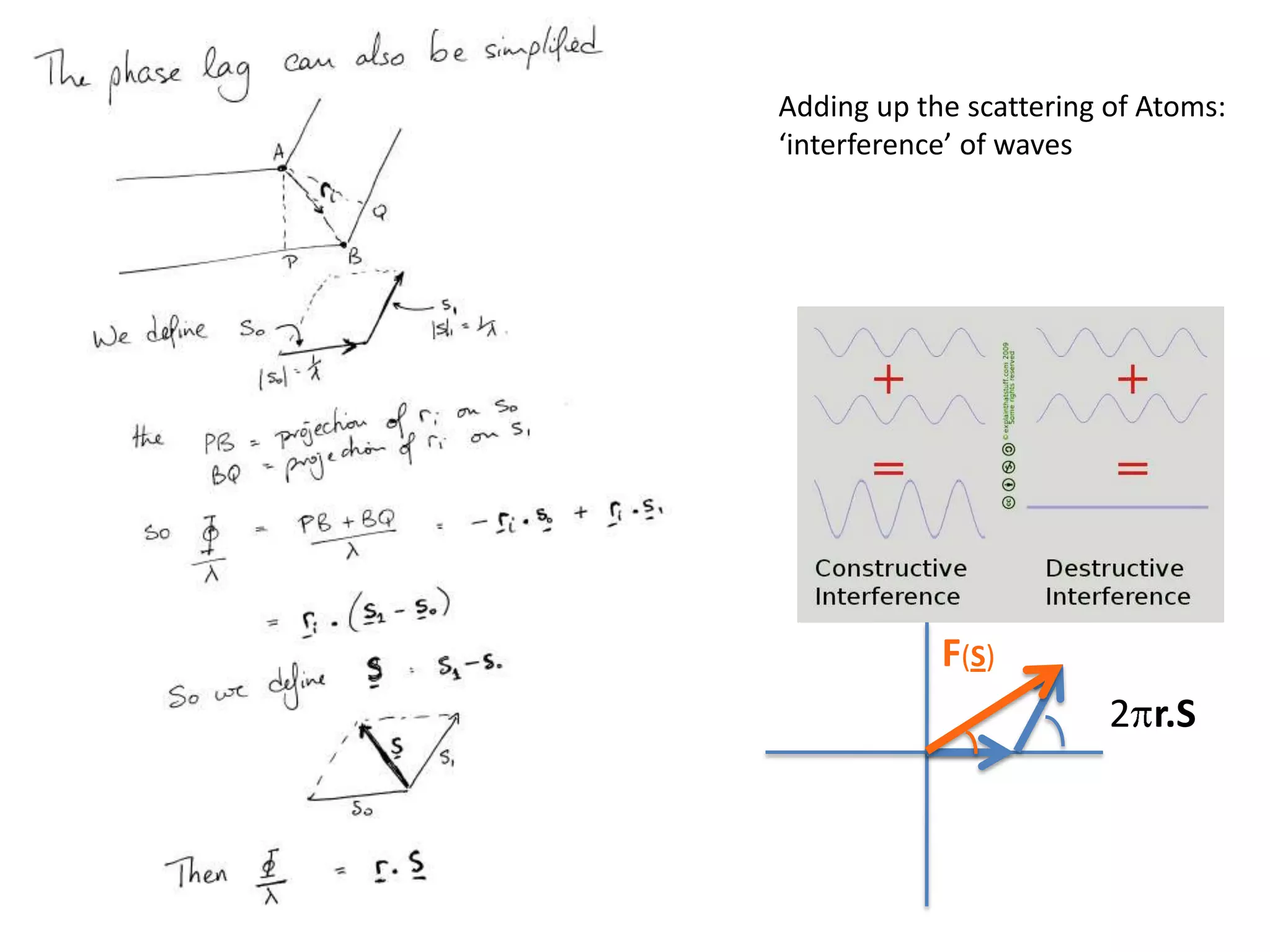
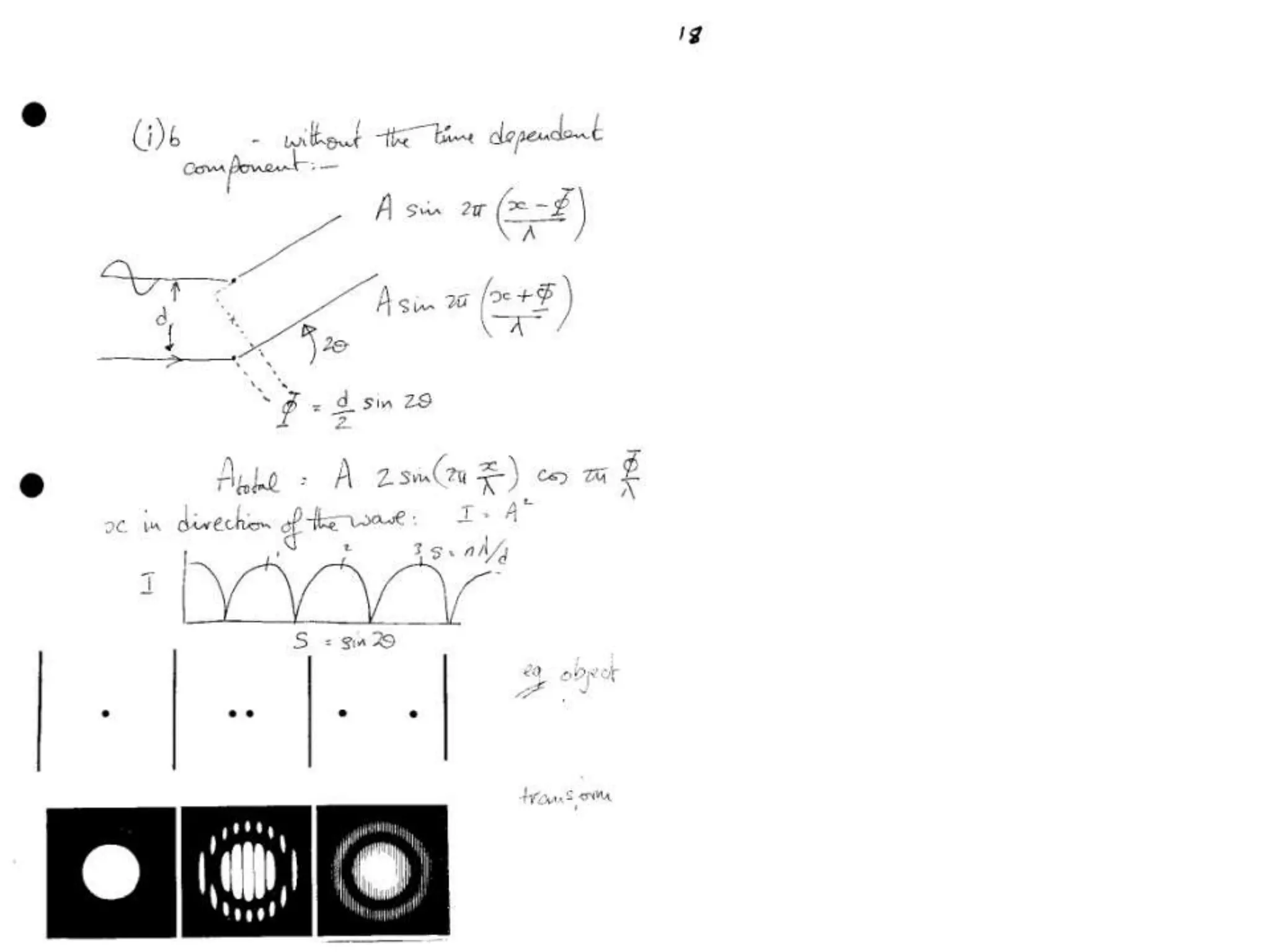
![ScatteringAdding up the scattering of Atoms:‘interference’ of wavesWaves add out of phaseby 2p[extra path/l]](https://image.slidesharecdn.com/bp219-class-4-04-2011-110411120321-phpapp01/75/BP219-class-4-04-2011-22-2048.jpg)
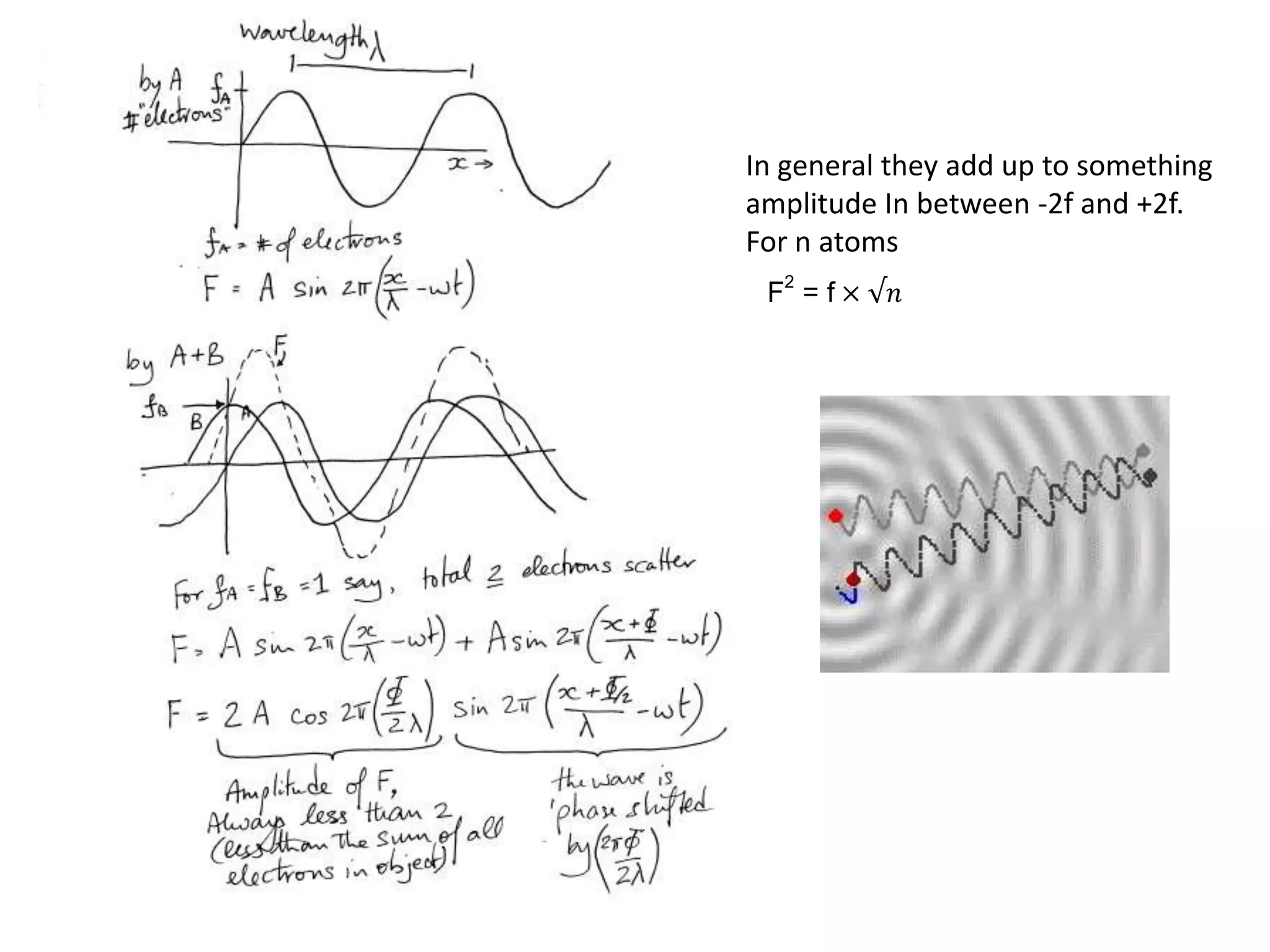
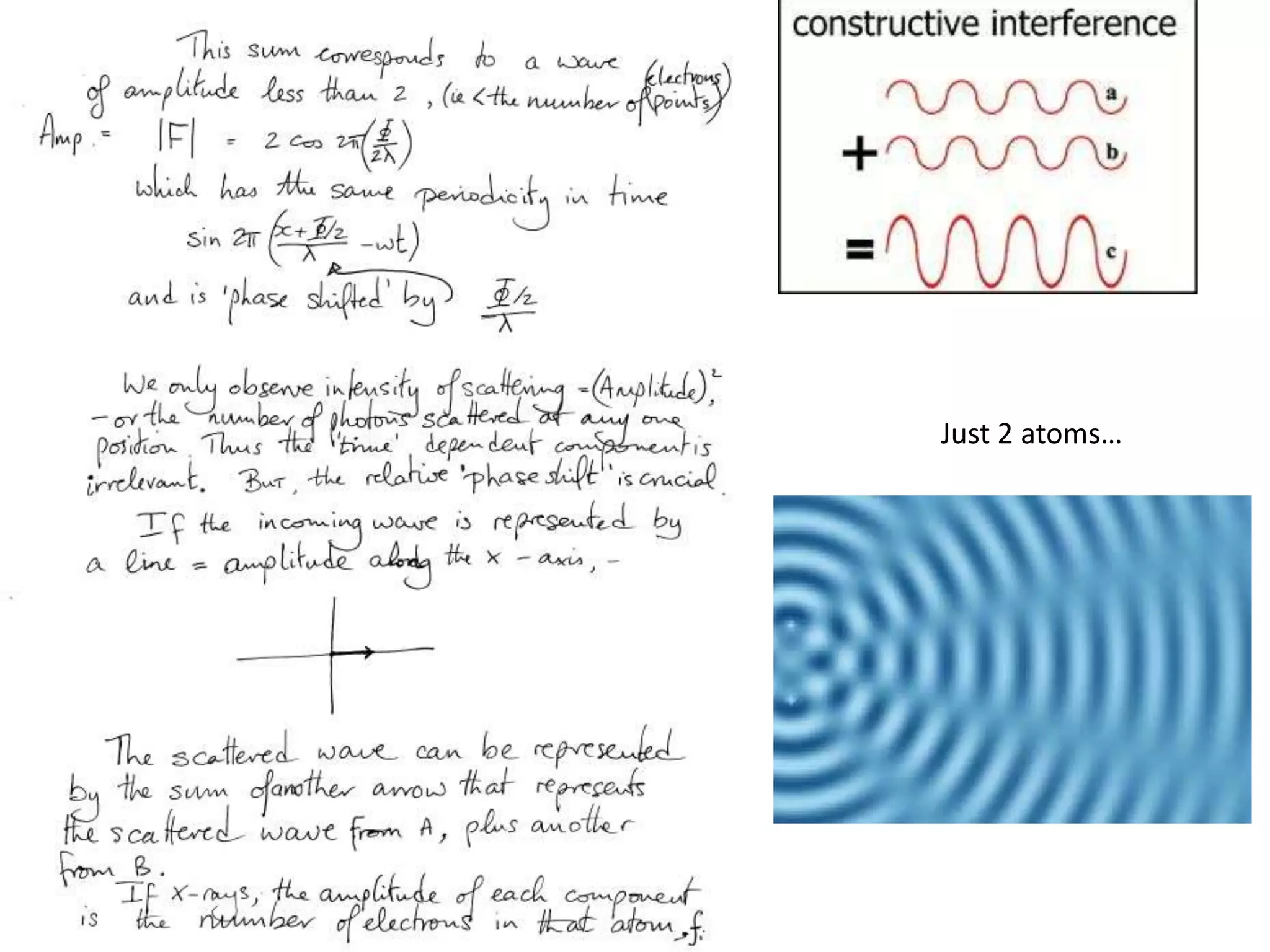
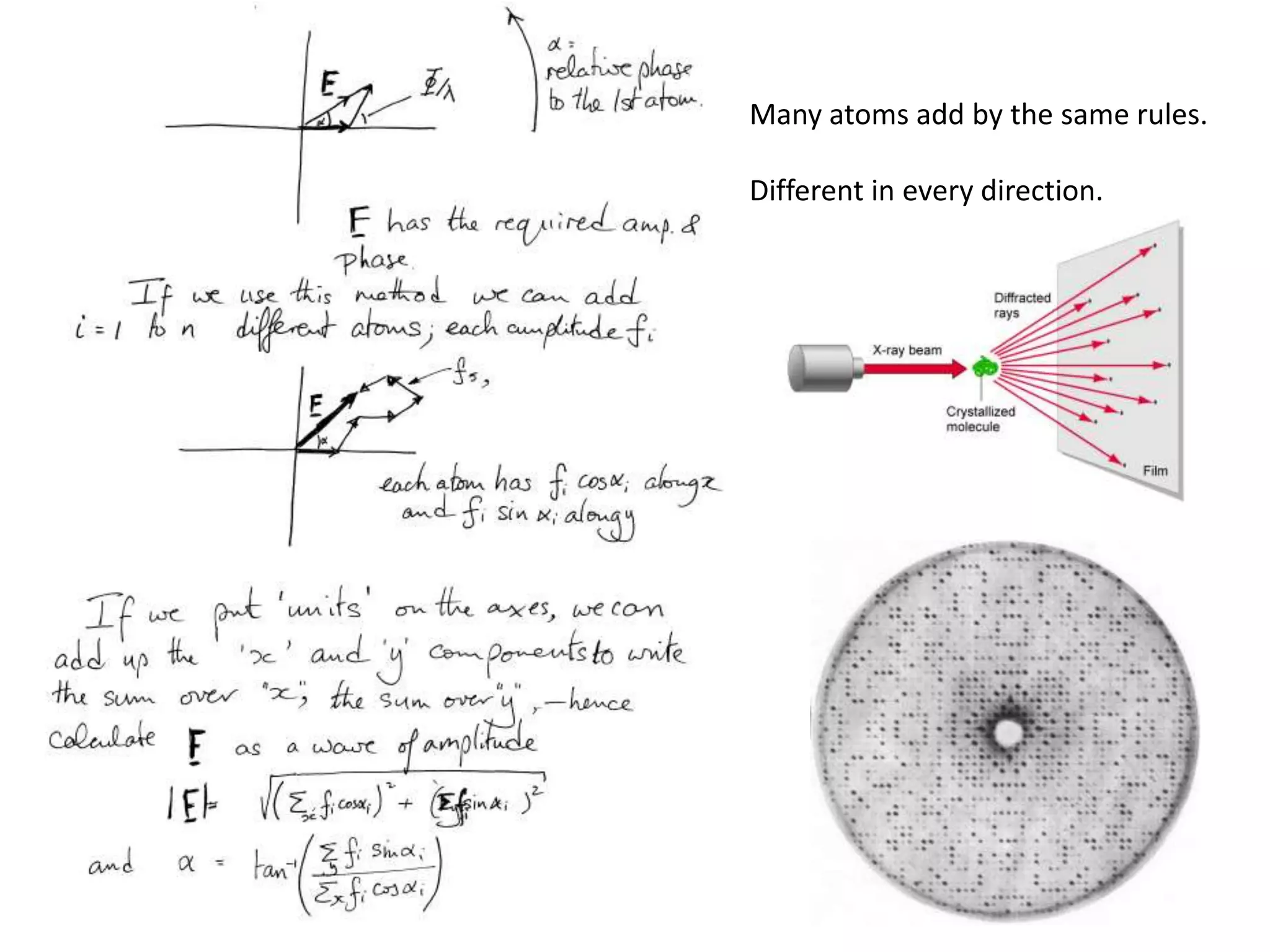
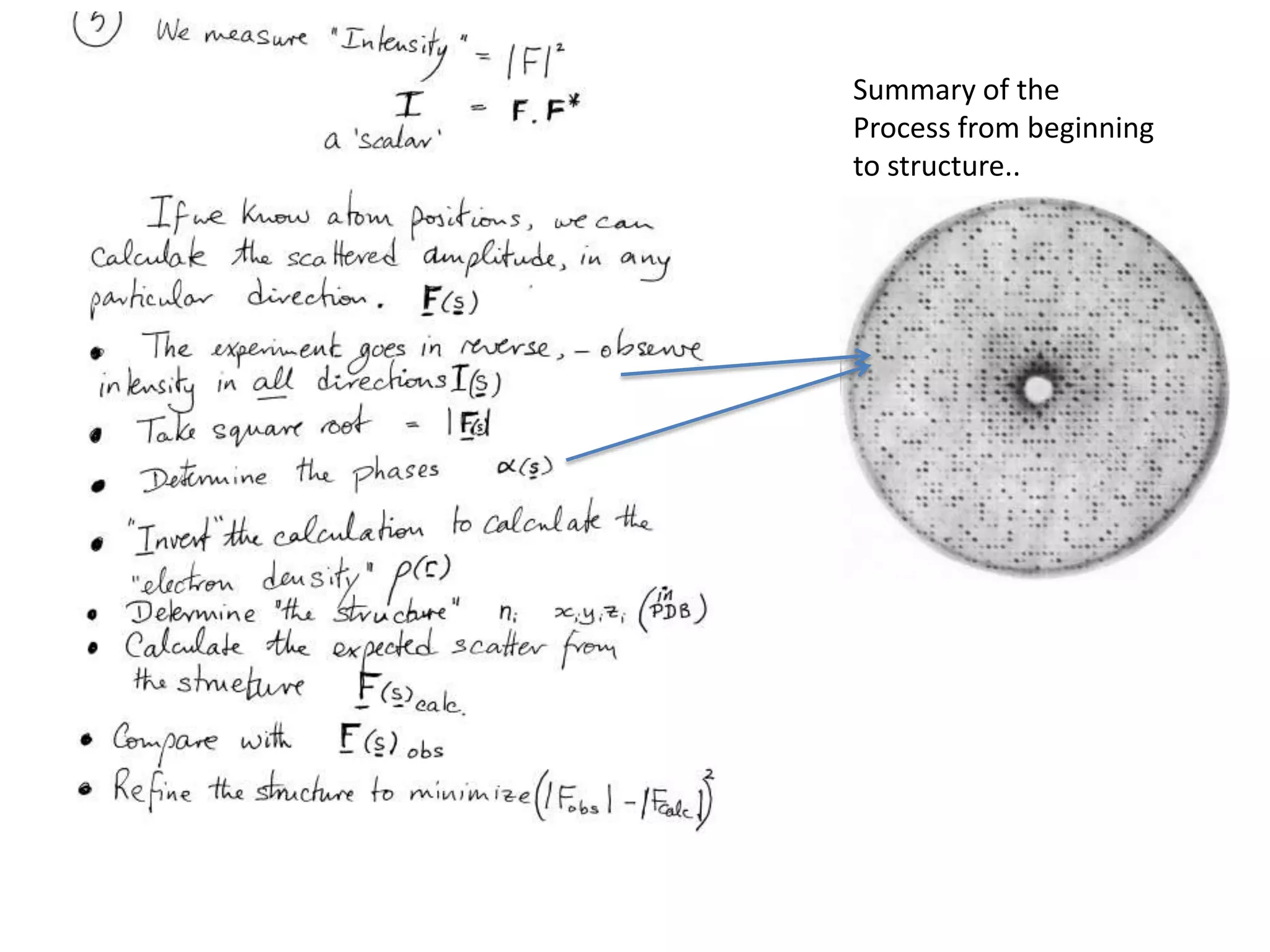


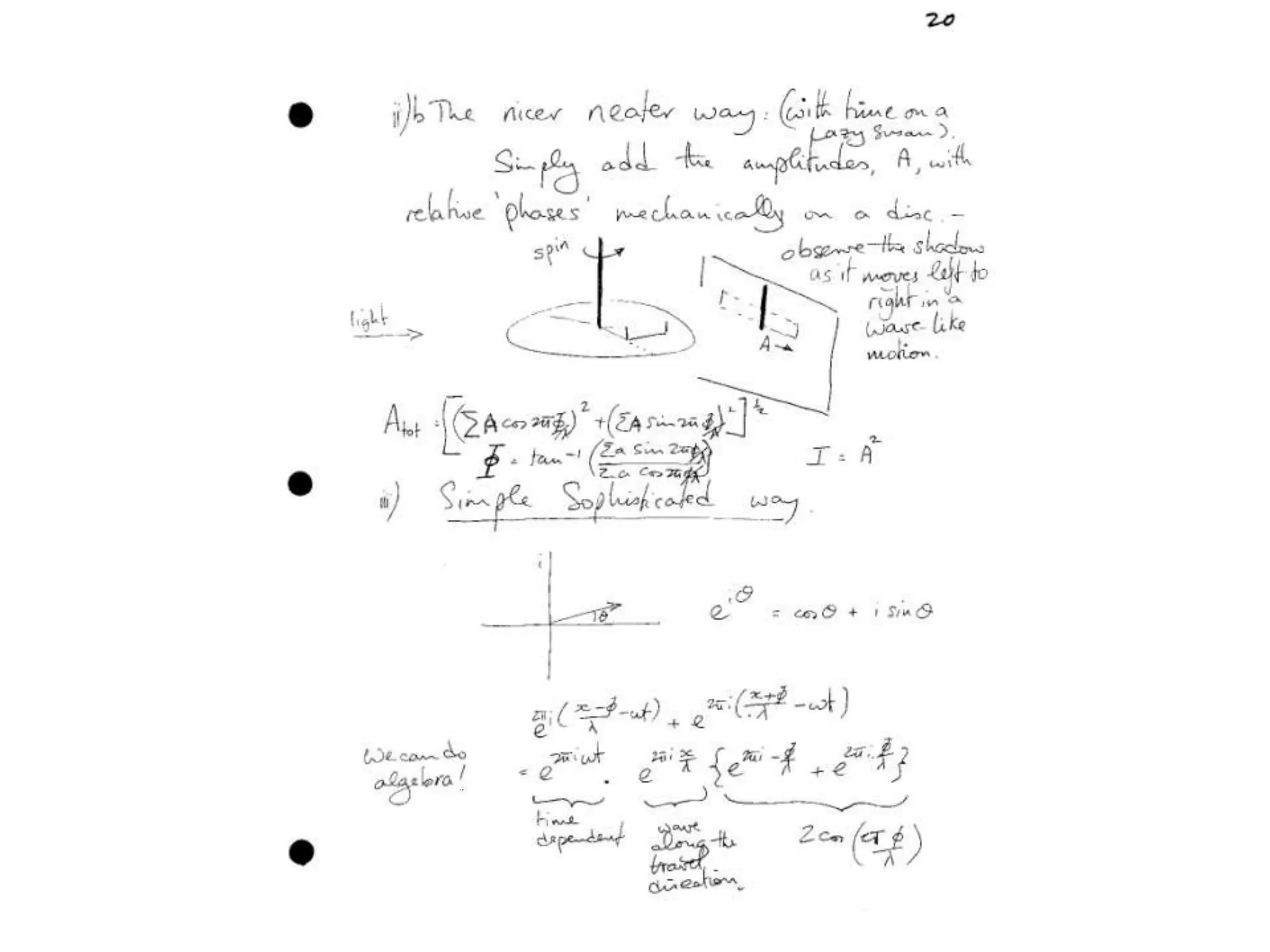
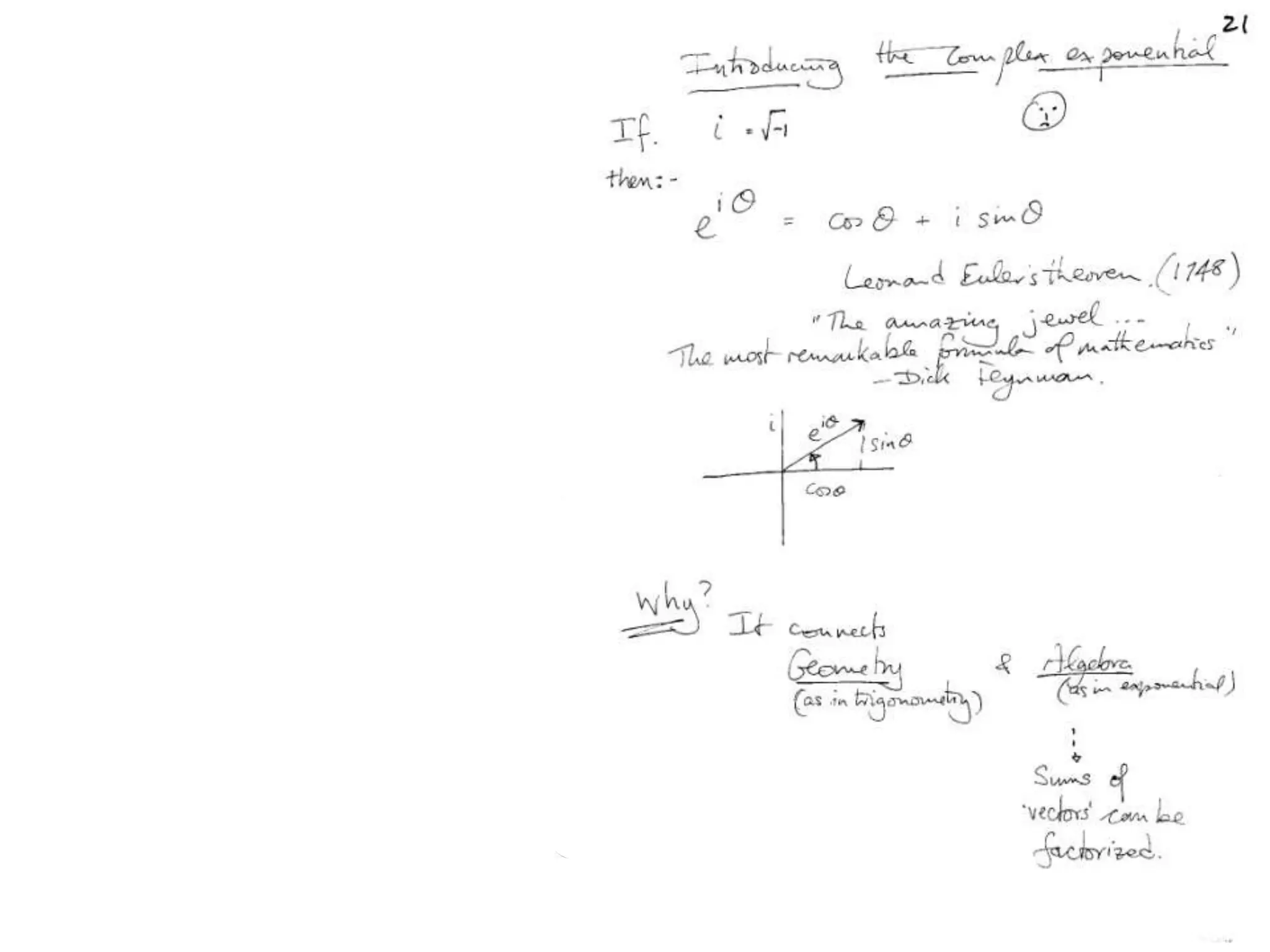


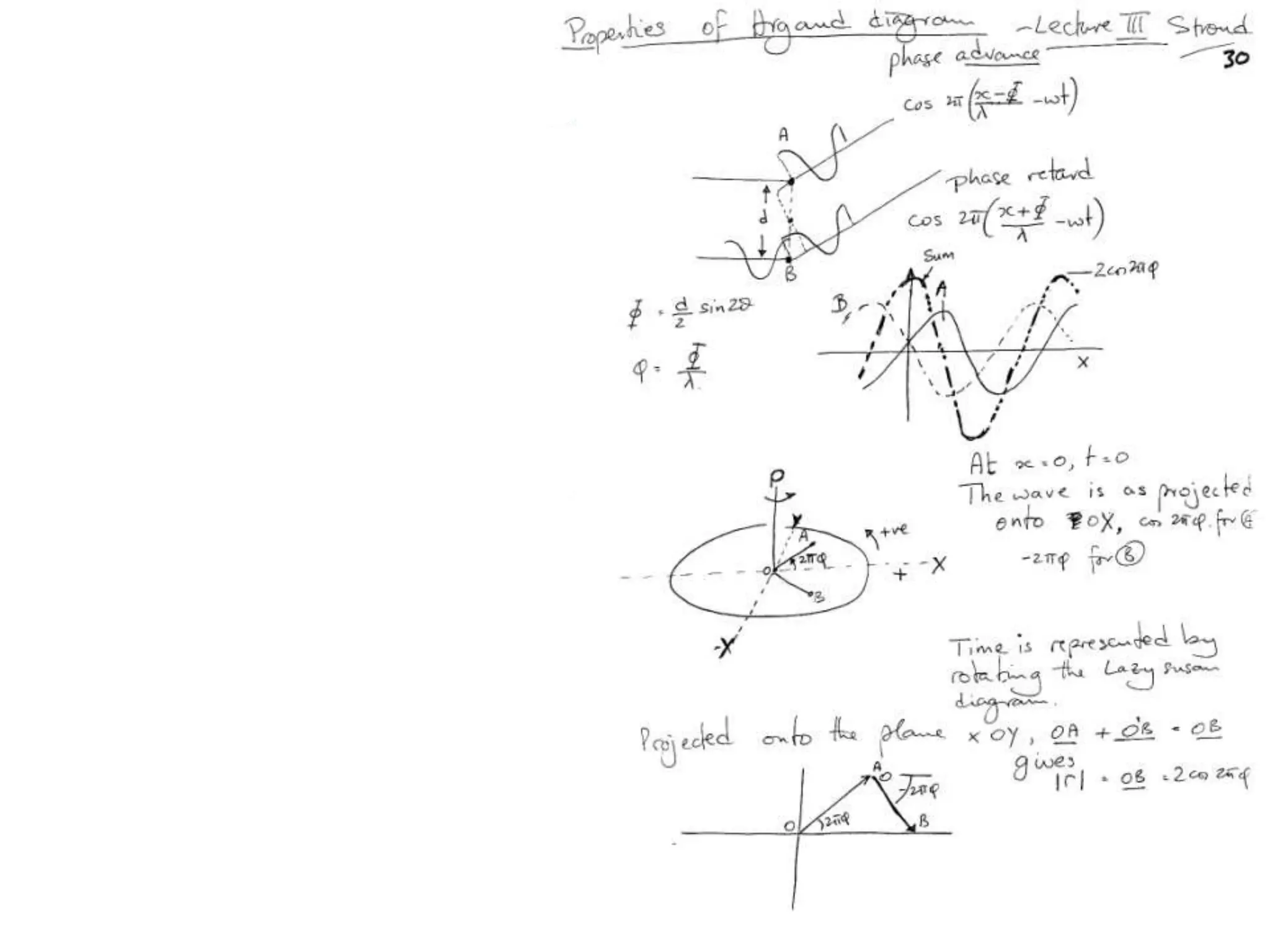



![Argand Diagram.. F(S) = |F(s)| eiqIntensity = |F(s)|2How to represent I(s)?|F(s)|2 =F(S) .F*(S) proof?Recall F*(S) is the complex conjugate of F(S) = |F(s)| e-iqso |F(s)|2 =|F(s)|[cos(q) + isin(q)].|F(s)|[cos(q) - isin(q)]=|F(s)|2 [cos2 (q) + sin2 (q)]=|F(s)|2 R.T.P.F(S)2pr.Sq](https://image.slidesharecdn.com/bp219-class-4-04-2011-110411120321-phpapp01/75/BP219-class-4-04-2011-45-2048.jpg)
![Argand Diagram.. F(S) = |F(s)| eiqIntensity = |F(s)|2How to represent I(s)?I(s) = |F(s)|2 =F(S) .F*(S) proof?Where F*(S) is defined to be the ‘complex conjugate’ of F(S) = |F(s)| e-iqso |F(s)|2 =|F(s)|[cos(q) + isin(q)].|F(s)|[cos(q) - isin(q)]=|F(s)|2 [cos2 (q) + sin2 (q)]=|F(s)|2 R.T.P.F(S)2pr.Sq](https://image.slidesharecdn.com/bp219-class-4-04-2011-110411120321-phpapp01/75/BP219-class-4-04-2011-46-2048.jpg)





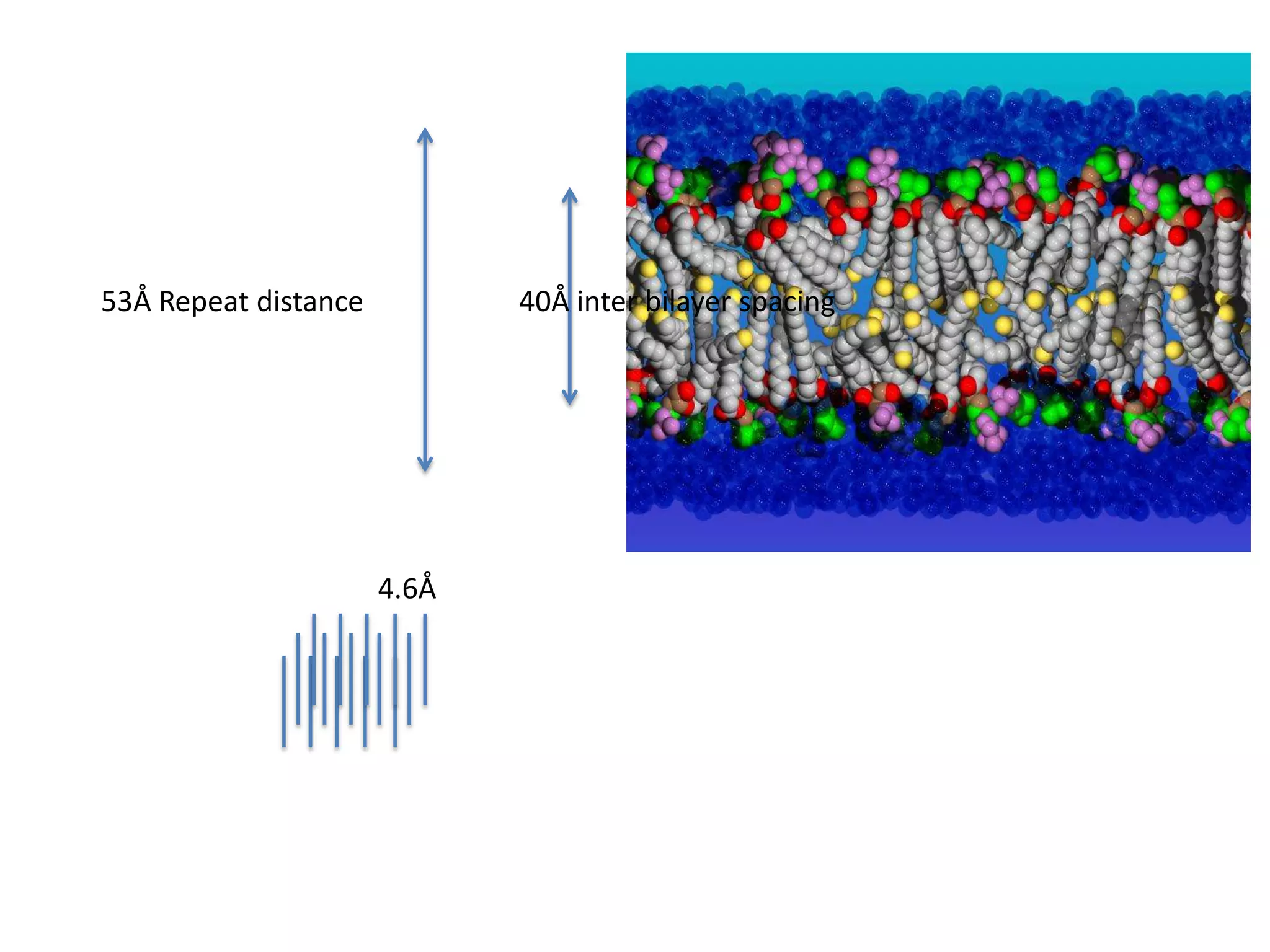
![Transform of two hoizontal lines defined y= ± y1F(s) = Int [x=0-infexp{2pi(xa+y1b).s} + exp{2pi(xa-y1b).s}dVr ]=2 cos (2py1b).s * Int [x=0-infexp{2pi(xa).s dVr]for a.s=0 the int[x=0-infexp{2pi(xa).s dVr] = total e content of the linefor a.s≠0 int[x=0-infexp{2pi(xa).s dVr] = 0hence r(r)isa line at a.s= 0 parallel to b, with F(s) = 2 cos (2py1b).s -along a vertical line perpendicular to the horizontal lines.Transform of a bilayer..b=53Å](https://image.slidesharecdn.com/bp219-class-4-04-2011-110411120321-phpapp01/75/BP219-class-4-04-2011-72-2048.jpg)