The document outlines the agenda for the 2016 Biophysics by the Sea conference, covering sessions on fluorescence spectroscopy, microscopy, and various biophysical applications. It includes detailed time slots for presentations by multiple researchers from various institutions, focusing on topics such as super-resolution imaging, molecular mechanics, and single-molecule spectroscopy. The conference also features discussions on innovative techniques and their implications in biophysics research.








![9
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Non-linear image scanning microscopy
1Ingo Gregor, 2Robert Ros, 1Jörg Enderlein
1III. Institute of Physics – Biophysics, Georg-August-University Göttingen
2Department of Physics and Center for Biological Physics, Arizona State University, Tempe AZ
ingo.gregor@phys.uni-goettingen.de, www.joerg-enderlein.de
Nowadays, multiphoton microscopy can be considered as a routine method for the
observation of living cells, organs, up to whole organisms. Second-harmonics generation (SHG)
imaging has evolved to a powerful qualitative and label-free method for studying fibrillar
structures, like collagen networks. However, examples of super-resolution non-linear
microscopy are rare. So far, such approaches require complex setups and advanced
synchronization of scanning elements limiting the image acquisition rates. We describe theory
and realization of a super-resolution image scanning microscope [1, 2] using two-photon
excited fluorescence as well as second-harmonic generation. It require only minor
modifications compared to a classical two-photon laser-scanning microscope and allows
image acquisition at the high frame rates of a resonant galvo-scanner. We achieve excellent
sensitivity and high frame-rate in combination with two-times improved lateral resolution. We
applied this method to fixed cells, collagen hydrogels, as well as living fly embryos. Further,
we verified the excellent image quality of our setup for deep tissue imaging.
[1] Müller C.B. and Enderlein J. (2010) Image scanning microscopy. Phys. Rev. Lett. 104(19), 198101.
[2] Sheppard C.J.R. (1988) Super-resolution in confocal imaging. Optik (Stuttg) 80 53–54.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-9-320.jpg)
![10
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Dead-time correction of fluorescence lifetime measurements
and fluorescence lifetime imaging
Sebastian Isbaner, Narain Karedla, Daja Ruhlandt, Simon Christoph Stein, Anna Chizhik,
Ingo Gregor, Jörg Enderlein
III. Institute of Physics – Biophysics, Georg August University, Göttingen
Dead-time artifacts can dramatically influence the shape of Time-Correlated Single Photon
Counting (TCSPC) histograms such as fluorescence lifetime curves [1]. These artifacts occur at
high count rates, which limit the acquisition speed in Fluorescence Lifetime Imaging
Microscopy (FLIM). We present an algorithm that corrects the distortions of TCSPC histograms
which are caused by constant electronics and/or detector dead-times [2]. We verified the
algorithm with Monte-Carlo simulations and fluorescence lifetime measurements.
Furthermore, we performed FLIM measurements on densely labeled cells at various excitation
powers and corrected the lifetime and intensity values for each pixel. Our correction method
is not restricted to TCSPC measurements only, but can be applied to any periodic single-event
counting or timing measurement. Since it corrects dead-time artifacts for both lifetime and
intensity, the algorithm could be beneficial for example for lidar or time-resolved fluorescence
anisotropy measurements.
[1] W. Becker, Advanced Time-Correlated Single Photon Counting Techniques (Springer, 2005).
[2] S. Isbaner, N. Karedla, D. Ruhlandt, S.C. Stein, A. Chizhik, I. Gregor, and J. Enderlein, Opt. Express 24, 9429-9445 (2016)](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-10-320.jpg)
![11
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Generation 3 Programmable Array Microscope (PAM) for
adaptive, high speed, large format optical sectioning
Donna J. Arndt-Jovin, Anthony H. B. de Vries, Thomas M. Jovin
Laboratory of Cellular Dynamics, Max-Planck-Institute for Biophysical Chemistry, Göttingen
djovin@mpibpc.mpg.de
We report on the current version of the optical sectioning programmable array
microscope(PAM) implemented with a digital micro-mirror device (DMD) as a spatial light
modulatorutilized for both fluorescence excitation and emission detection. The PAM is based
on structured illumination [1]. A sequence of HD (1920×1080) binary patterns of excitation
light is projected into the focal plane of the microscope at the 18 kHz binary frame rate of the
TI1080p DMD. The resulting sequence of patterned emissions is captured in a single
acquisition as two distinct images: conjugate (ca. “on-focus”) consisting of signals impinging
on and deviated from the “on” elements of the DMD, and the non-conjugate (ca. “out-of-
focus”) of those falling on and deviated from the “off” elements. The sectioned image is gained
from a weighted subtraction of the conjugate and non-conjugate images. This procedure
allows for a high duty cycle (typically 30 to 50%) of on-elements in the excitation patterns and
thus functions well with low light intensities, preventing saturation of the fluorophores. The
corresponding acquisition speed is also very high, limited only by the bandwidth of the
camera(s) (100 fps full frame with the current sCMOS camera) and the optical power of the
light source (lasers, LEDS). In contrast to the static patterns typical of SIM systems, the
programmable array allows optimization of the patterns to the sample (duty cycle and feature
size), as well as enabling a wide range of microscopy applications, ranging from patterned
photobleaching, (FRAP, FLIP) and photoactivation, spatial superresolution (SIM, etc.),
automated adaptive minimized light exposure (MLE) [2], and photolithography. This work is
supported by BMBF VIP Grant 03V0441 (iPAM: "Intelligentes" Programmierbares Array
Mikroskop).
[1] de Vries, A., N. Cook, S. Kramer, D. Arndt-Jovin and T. Jovin (2015). "Generation 3 programmable array microscope (PAM) for high
speed, large format optical sectioning in fluorescence." Proc. SPIE 9376(93760C): 1-15
[2] W. Caarls; B. Rieger, A.H.B. de Vries, D.J. Arndt-Jovin, T.M. Jovin (2010). “Minimizing light exposure with the programmable array
microscope”, J. MICROSCOPY, 241, 101-110](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-11-320.jpg)




![17
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Surface motility and colony growth in bacteria
Stefan Klumpp
Institute for Nonlinear Dynamics, Georg-August- University Göttingen
Max-Planck-Institute of Colloids and Interfaces, Potsdam
Motile bacteria move through a variety of mechanisms, which employ different molecular
machines. Often, physical forces play a key role. I will discuss this using the role of mechanical
interactions in twitching motility as an example. Twitching motility is a mode of motion on
surfaces that is driven by the retraction of type IV pili, filamentous appendages that pull the
cell forward through cycles of growth, attachment to the surface and retraction into the cell,
driven by APTases at the base of the pili. In some bacterial species multiple pili pull the cell in
different directions simultaneously. Thus, the pili perform a two-dimensional tug-of-war.
Tugof- war-like interactions, where molecular motors exert forces on each other, were
previously studied for bidirectional cytoskeletal transport. I will review this case, which is one-
dimensional and show that the tug-of-war provides a mechanism for persistent directionality.
In the two-dimensional case, the tug-of-war is less efficient at doing so than in one dimension,
as will be shown for the case of the twitching motility of N. gonorrhoeae, where an additional
mechanisms for directional memory was predicted theoretically and confirmed
experimentally [1]. N. gonorrhoeae bacteria use twitching to find each other in order to
initiate the formation of colonies. As a second topic, I will discuss the growth of planar colonies
and its interplay with the adhesion between cells that is also mediated by the type IV pili. To
that end, a minimal model for mixed colonies of cells of different adhesion is presented [2].
The model effectively combines differential adheision with rangeexpansion-like growth.
[1] R. Marathe, C. Meel, …, B. Meier, S. Klumpp, Nature Comm. 5, 3759 (2014)
[2] J.J. Dong and S. Klumpp, unpublished](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-17-320.jpg)
![18
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Mechano-Sensitivity is Cell Type Specific
Galina Kudryasheva, Florian Rehfeldt
III. Institute of Physics – Biophysics, Georg-August- University Göttingen
galina.kudryasheva@phys.uni-goettingen.de
Nowadays it is widely acknowledged that cellular fate is dependent on the mechanical
properties of their micro-environment. Cells sense the stiffness of their surrounding with
contractile acto-myosin stress fibers through focal adhesions and react to such physical stimuli
by altering their bio-chemical pathways. Human mesenchymal stem cells (hMSCs) are an
especially striking as their differentiation towards various cell types can be guided not only by
chemical induction, but also by tuning the extracellular matrix stiffness. While the entire
differentiation process can take several days up to weeks, the structure and dynamics of stress
fibers can be used as an early morphological marker and theoretically modelled using classical
mechanics with an active spring model [1]. We use this approach to analyze the mechanical
cell-matrix interactions of hMSCs and several types of differentiated cells.
We plated cells on elastic poly-acrylamide hydrogels covering the whole physiological range
of stiffness given by Young’s moduli E from 1 to 130 kPa. Using immunofluorescence we
visualized stress fibers and analyzed the cytoskeletal morphology [2]. Analyzing cell area and
cytoskeletal order parameter we could assign an effective cellular stiffness that shows
distinct differences during the differentiation process and for different cell types. Our
experiments show that cellular susceptibility to the substrate elasticity is highly cell type
specific and dependent on acto-myosin contractility.
[1] A. Zemel et al. Nat.Phys. 6, 468–473 (2010)
[2] B. Eltzner et al. PLoS One 10 (2015)](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-18-320.jpg)





![24
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
New analysis method for passive microrheology
1Kengo Nishi, 2Maria L. Kilfoil, 1Christoph F. Schmidt, 3Fred C. MacKintosh
1III. Institute of Physics - Biophysics, Georg August University Göttingen
2Univ. of Massachusetts, Amherst, MA
3Department of Physics & Astronomy, Faculty of Sciences, Vrije Universiteit Amsterdam
kengo.nishi@phys.uni-goettingen.de
Passive microrheology is an experimental technique used to measure the mechanical
response of materials from the fluctuations of micron-sized beads embedded in the medium.
Microrheology is well suited to study rheological properties of materials that are difficult to
obtain in larger amounts and also of materials inside of single cells. In one common approach,
one uses the fluctuation-dissipation theorem to obtain the imaginary part of the material
response function from the power spectral density of bead displacement fluctuations, while
the real part of the response function is calculated using a Kramers-Kronig integral. The high-
frequency cut-off of this integral strongly affects the real part of the response function in the
high frequency region. Here, we discuss how to obtain more accurate values of the real part
of the response function by an alternative method using autocorrelation functions.
[1] B. Schnurr, F. Gittes, F. C. MacKintosh, and C. F. Schmidt, Macromolecules, 1997, 70, 7781-7792.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-24-320.jpg)
![25
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Mechanics Matters for Cells: Forces, Elasticity, and Cytoskeleton
Florian Rehfeldt
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
rehfeldt@physik3.gwdg.de, www.florian-rehfeldt.de
The mechanical properties of microenvironments in our body vary over a broad range and are
as important to cells as traditional biochemical cues. An especially striking experiment of this
mechano-sensitivity demonstrated that systematic variation of the Young’s elastic modulus E
of the substrate can direct the lineage differentiation of human mesenchymal stem cells
(hMSCs) (1).
To elucidate the complex interplay of physical and biochemical mechanisms of cellular
mechano-sensing, well-defined extracellular matrix (ECM) models are essential. While elastic
substrates made of poly-acrylamide (PA) are widely in use, they have the potential drawback
that the precursors are cytotoxic and therefore do not allow for 3D culture systems. Here, a
novel biomimetic ECM model based on hyaluronic acid (HA) was successfully established that
exhibits a widely tuneable and well-defined elasticity E, enables 2D and 3D cell culture and
enables us to mimic a variety of distinct in vivo microenvironments (2). Quantitative analysis
of the structure of acto-myosin fibers of hMSCs on elastic substrates with an order
parameter S, reveals that the stress fiber morphology is an early morphological marker of
mechano-guided differentiation and can be understood using a classical mechanics model (3-
5). Furthermore, the cytoskeleton also dictates the shape of the nucleus and lends support to
a direct mechanical matrix-myosin-nucleus pathway (6).
[1] Engler, A. J., S. Sen, H. L. Sweeney, and D. E. Discher. 2006. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 126:677-
689.
[2] Rehfeldt, F., A. E. X. Brown, M. Raab, S. Cai, A. L. Zajac, A. Zemel, and D. E. Discher. 2012. Hyaluronic acid matrices show matrix
stiffness in 2D and 3D dictates cytoskeletal order and myosin-II phosphorylation within stem cells. Integrative Biology 4:422-430.
[3] Zemel, A., F. Rehfeldt, A. E. X. Brown, D. E. Discher, and S. A. Safran. 2010. Optimal matrix rigidity for stress-fibre polarization in
stem cells. Nature Physics 6:468-473.
[4] Zemel, A., F. Rehfeldt, A. E. X. Brown, D. E. Discher, and S. A. Safran. 2010. Cell shape, spreading symmetry, and the polarization
of stress-fibers in cells. J Phys-Condens Mat 22.
[5] Paluch, E. K., C. M. Nelson, N. Biais, B. Fabry, J. Moeller, B. L. Pruitt, C. Wollnik, G. Kudryasheva, F. Rehfeldt, and W. Federle. 2015.
Mechanotransduction: use the force(s). BMC Biology 13:1-14.
[6] Swift, J., I. L. Ivanovska, A. Buxboim, T. Harada, P. C. D. P. Dingal, J. Pinter, J. D. Pajerowski, K. R. Spinler, J.-W. Shin, and M. Tewari.
2013. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 341.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-25-320.jpg)
![26
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Osmosis and force fluctuation of non-adhering cells
1Christoph F. Schmidt, 2Todd M. Squire, 1Samaneh Rezvani
1 III. Institute of Physics – Biophysics, Georg-August-University Göttingen
2Department of Chemical Engineering, University of California. Santa Barbara, CA
srezvani@physik3.gwdg.de
Cells sense their micro-environment through biochemical and mechanical interactions. They
can respond to stimuli by undergoing shape- and possibly volume changes. Key components
in determining the mechanical response of a cell are the viscoelastic properties of the
actomyosin cortex, effective surface tension, and the osmotic pressure. We use custom-
designed microfluidic chambers with integrated hydrogel micro windows to be able to rapidly
change solution conditions for cells without any hydrodynamic flow. We use biochemical
inhibitors and different osmolytes and investigate the immediate response of individual cells.
Using a dual optical trap makes it possible to probe suspended rounded-up cells by active and
passive microrheology to quantify the response to the various stimuli.
[1] F. Schlosser, F. Rehfeldt and C. F. Schmidt, Phil. Trans. R. Soc. B 370, 0028 (2014)
[2] Joel S. Paustian and Todd M. Squires, Phys Rev 3, 041010 (2013)](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-26-320.jpg)





![32
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Are protein hydration and dynamics important factors in the enzyme kinetics? –
fluorescence study on Haloalkane-dehalogenases
1Jan Sýkora, 2Jan Brezovský, 1Mariana Amaro, 3Silvia Kováčová, 1Avisek Ghose,
2Zbyněk Prokop, 2Koen Beerens, 2Šárka Bidmanová, 2Radka Chaloupková, 3Kamil Paruch,
2Jiří Damborský, 1Martin Hof
1Department of Biophysical Chemistry, J. Heyrovsky Institute of Physical Chemistry,
Czech Academy of Sciences, Prague
2Loschmidt Laboratories, Department of Experimental Biology and Research Centre for Toxic
Compounds in the Environment RECETOX, Faculty of Science, Masaryk University, Brno
3Department of Chemistry, Faculty of Science, Masaryk University, Brno
jan.sykora@jh-inst.cas.cz
The hydration and mobility of proteins are believed to profoundly affect their function1.
However, only a few approaches for monitoring these characteristics within the relevant
protein regions are available. Here we describe two general methods for site-specific analysis
of the extent of hydration and degree of the mobility in enzyme Haloalkane Dehalogenase.
The first approach is based on recording „time dependent fluorescence shift“ (TDFS)2 placing
the dye in the tunnel mouth of this enzyme3,4. In the latter approach, environment sensitive
coumarin dye is inserted in the selected region employing the technology of the “unnatural
aminoacid”5. By means of the steady state spectroscopy the degree of hydration can be
determined including the presence of ‘structured water’6. Finally, the „gating“ dynamics of
the enzymes can be traced by following the photoinduced electron transfer (PET) between
the selected tryprophan and properly positioned fluorescence dye7. Both the hydration and
dynamics monitored within the biologically relevant regions of the dehalogenase enzymes is
then compared with their enzyme kinetics of various mutants, which can bring the deeper
insight into the functioning of these enzymes.
[1] Levy, Y.; Onuchic, J. N. Annu. Rev. Biophys. Biomolec. Struct. 2006, 35, 389.
[2] Horng, M. L. et al. J. Phys. Chem. 1995, 99, 17311.
[3] Amaro, M. et al. J. Phys. Chem. B 2013, 117, 7898.
[4] Sykora, J. et al. J. Nat. Chem. Biol. 2014, 10, 428.
[5] Summerer, D. et al. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 9785.
[6] Amaro, M. et al. J. Am. Chem. Soc. 2015, 137, 4988.
[7] Sauer, M.; Neuweiler, H. In Fluorescence Spectroscopy and Microscopy; Engelborghs, Y., Visser, A. J. W. G., Eds.; Humana Press: 2014;
Vol. 1076, p 597.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-32-320.jpg)
![33
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Self-organization of computation in neural systems by interaction between
homeostatic and synaptic plasticity
Christian Tetzlaff
III. Institute of Physics - Biophysics, Georg-August-University Göttingen
Bernstein Center for Computational Neuroscience, Georg-August-University Göttingen
Max-Planck Institute for Dynamics and Self-Organization, Göttingen
tetzlaff@phys.uni-goettingen.de
The ability to perform complex motor control tasks is essentially enabled by the nervous
system via the self-organization of large groups of neurons into coherent dynamic activity
patterns. During learning, this is brought about by synaptic plasticity, resulting in the
formation of multiple functional networks – commonly termed as ‘cell-assemblies’. A
multitude of such cell assemblies provide the requisite machinery for non-linear computations
needed for the mastery of a large number of motor skills. However, given the fact that there
exists considerable overlap between the usage of the same neurons within such assemblies,
for a wide range of motor tasks, creation and sustenance of such computationally powerful
networks posses a challenging problem. How such interwoven assembly networks self-
organize and how powerful assemblies can coexist therein, without catastrophically
interfering with each other remains largely unknown. On the one side, it is already known that
networks can be trained to perform complex nonlinear calculations [1], such that, if the
network possesses a reservoir of rich, transient dynamics, desired outputs can be extracted
from these reservoirs in order to enable motor control. On the other side, cell assemblies are
created by hebbian learning rules that strengthen a synapse if pre- and post-synaptic neurons
are co-active within a small enough time window [2]. Therefore, it appears relatively
straightforward to combine these mechanisms in order to construct powerful assembly
networks. However, given that the self-organization of neurons into cell assemblies by the
processes of synaptic plasticity induces ordered or synchronized neuronal dynamics, which
can destroy the required complexity of a reservoir network, such a combination remains a
very challenging problem [3]. Furthermore, simultaneous creation of multiple cell assemblies
can also lead to catastrophic interference if one cannot prevent them from growing into each
other. In this study, we exploit for the first time the interaction between neuronal and synaptic
processes acting on different time scales to enable, on a slow timescale, the self-organized
formation of assembly networks (Fig. 1), while on a faster timescale, to conjointly perform
several non-linear calculations needed for motor fine-control. Specifically, by the combination](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-33-320.jpg)
![34
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
of synaptic plasticity and synaptic scaling [4], as a homeostatic mechanism, we demonstrate
that such self-organization allows executing a difficult, six degrees of freedom, manipulation
task with a robot where assemblies need to learn computing complex non-linear transforms
and - for execution - must cooperate with each other without interference. This mechanism,
thus, permits for the first time, the guided self-organization of computationally powerful sub-
structures in dynamic networks for behavior control. Furthermore, comparing our assembly
network to networks with unchanging synapses ("static" networks) shows that it is indeed the
embedding of a strongly connected assembly that creates the necessary computational
power.
[1] Buonomano DV, Maass W. Nat. Rev. Neurosci 2009, 10:113-125.
[2] Palm, G. et al. Biol. Cybern., 108:559 -572, 2014.
[3] Klamp, S. and Maass, W. J. Neurosci., 33(28):11515 11529, 2013.
[4] Tetzlaff, C. et al. PLoS Comput. Biol., 9(10):e10003307, 2013.
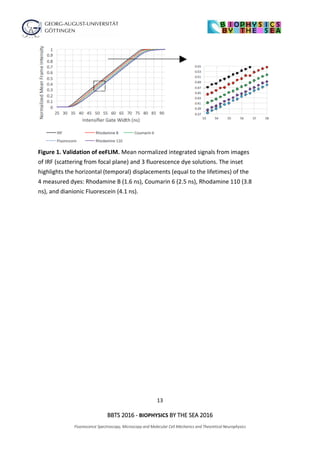
Figure 1: Cell assembly size and
computational performance are correlated.
(A) Input driven formation of cell assemblies
brought about by the interaction long-term
potentiation (LTP) and synaptic scaling (Syn.
Sca.). (B) With more learning trials the
assembly grows and integrates more
neurons. We measure this by arbitrarily
defining assembly size by that set of neurons
connected with efficacies larger than half
the maximum weights. (C) Parallel to the
outgrowth of the cell assembly the error of the
system to perform several linear and non-linear
calculations decreases.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-34-320.jpg)


![37
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Enhancing the performance and applicability of SOFI using
new probes and analysis strategies
Wim Vandenberg, Sam Duwé, Peter Dedecker
Department of Chemistry, Katholieke Universiteit Leuven
wim.vandenberg@chem.kuleuven.be, www.chem.kuleuven.be/pd
In the past decade, one after the other, new ways of achieving super-resolution have been
thought up and implemented, targeting different niche parts of the imaging field. One of these
techniques, superresolution optical fluctuation imaging or SOFI (1) is targeting an audience
concerned with the robustness of the analysis (2). As such it’s truly in high background low-
signal situations (such as living systems) that SOFI comes in to its own. The technique is based
on a statistical analysis of several hundred images taken of a sample in which the label shows
fluorescence dynamics (blinking), the precise nature of this blinking is often irrelevant making
many different labels suitable (3,4). In the last couple of years SOFI has matured to deliver
multi-color (4) as well as 3D (5) imaging in living cells. In this contribution we will describe a
continuing focus on our part to quantify and enhance the robustness of SOFI in live cells. On
the one hand this work has focused on the development of fluorescent proteins with
increased bio-compatibility and good performance in SOFI microscopy (6). On the other hand
this work has focused on the development of a statistical framework which allows for the
model free quantification of the quality of SOFI datasets as well as an enhancement of the
SOFI analysis, allowing for the doubling of temporal resolution by using all available
information (7).
[1] Dertinger et al., “Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI)”
[2] Geissbuehler at al., "Comparison between SOFI and STORM"
[3] Dertinger et al., “Superresolution Optical Fluctuation Imaging with Organic Dyes”
[4] Dedecker et al., “Widely accessible method for superresolution fluorescence imaging of living systems”
[5] Geissbuehler at al. Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging”
[6] Duwé et al., “Expression-Enhanced Fluorescent Proteins Based on Enhanced Green Fluorescent Protein for Super-resolution
Microscopy”
[7] Vandenberg et al., “Model-free uncertainty estimation in stochastical optical fluctuation imaging (SOFI) leads to a doubled temporal
resolution”](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-37-320.jpg)

![39
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Expansion Microscopy meets dSTORM
1Fabian Zwettler, 1Felix Rüdinger, 1Markus Sauer
1Department of Biotechnology & Biophysics, Julius-Maximilians-University Würzburg
fabian.zwettler@uni-wuerzburg.de
Single molecule localization microscopy (SMLM) and the recently developed technique
Expansion Microscopy (ExM) 1 are two different approaches that achieve the visualization and
investigation of proteins and other biological molecules with nanoscale precision. SMLM
techniques such as direct stochastic optical reconstruction microscopy (dSTORM) bypasses the
diffraction limit of light microscopy by photoswitching or –activation of a sparse subset of all
fluorophores, localization of single molecules by fitting a two dimensional Gaussian function
to the photon distribution (PSF) of single fluorophores, and reconstruction of a super-resolved
image. In contrast to this technique, ExM increases the effective resolution through physically
magnifying the specimen. Therefore the specimen is embedded in a dense swellable polymer
in which a modified fluorescent tag is targeted to a biomolecule of interest. Additionally this
label is anchored into the polymer mesh. By adding water the polymer expands isotropically
in all dimensions and enables a 4.5x magnification of the specimen. This process improves the
spatial resolution down to roughly 60-70 nm in lateral direction on a diffraction-limited
microscope. By combining dSTORM with Expansion Microscopy we are able to further improve
the spatial resolution to molecular dimensions. Our new approach is a highly promising tool
that can be used advantageously to investigate the 3D molecular architecture of biomolecular
complexes and machines.
[1] Chen, F. Tillberg, P. W. & Boyden, E. S. Expansion microscopy. SCIENCE 347, 543–548 (2015).](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-39-320.jpg)

![41
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Self-Organized Memory Allocation by Hebbian Cell Assemblies
Johannes M. Auth, Timo Nachstedt, Christian Tetzlaff
III. Institute of Physics – Biophysics, Georg August University Göttingen
Bernstein Center for Computational Neuroscience, Göttingen, 37077, Germany
Max Planck Institute for Dynamics and Self-Organization, Göttingen, 37077, Germany
jauth@phys.uni-goettingen.de
Declarative memory denotes the storage of facts and concepts from perceived stimuli. The
formation of such memories, in particular their allocation in neural circuits is still an
unresolved problem. In general, different stimuli to be learned have to trigger the formation
of different memory representations. In addition, each learned stimulus has to maintain its
assignment or allocation to its specifically formed memory representation. Experimental
findings imply that variations in neural excitability due to a complex cascade of proteins that
make individual neurons more susceptible form a memory representation of a new stimulus
[1]. Furthermore, the concept of synaptic tagging, which assumes cascades of plasticity-
related proteins, is assumed to locally determine the synapses involved in the memorization
process [2]. However, both ideas require complex, highly specialized cascades of several
proteins to allocate memories. Here, we show in a theoretical model that the allocation of
memory can already be solved by the self-organized dynamics of synaptic plasticity. The
system consists of three neuronal populations: an input population projects activity patterns
(stimuli) through random excitatory connections on a second, recurrently interconnected
memory population. All feed-forward as well as the recurrent synapses are adapted by a
combination of Hebbian synaptic plasticity and synaptic scaling [3]. An inhibitory population
is mutually connected to the recurrent layer to provide global competition. Interestingly, first
of all, our model successfully forms stable memory representations: presenting a given
stimulus to the recurrent layer causes a locally clustered group of neurons to become strongly
interconnected with each other (Hebbian cell assembly [4]). Furthermore, presenting another
stimulus of sufficient dissimilarity to the first one causes the formation of another memory
representation. Remarkably, if the stimuli are quite similar to each other, both are allocated
to the same memory representation. In addition, the system shows the dynamics of
competitive memory recall, i.e. differentiating recognition [5], as the activation of one
memory representation fully suppresses others. In summary, the here-presented simple but
biologically plausible concepts of stimulus-dependent self-organization of plasticity provide a
promising approach to the question of how memory allocation is coordinated in the brain.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-41-320.jpg)
![42
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
[1] Yiu A. P. et al. Neuron 2014, 83(3): 722-735.
[2] Rogerson T. et al. Nat Rev Neuroscience 2014, 15(3): 157-169.
[3] Tetzlaff C. et al. PLoS Comput Biol 2013, 9(10):e1003307.
[4] Hebb, D. O.: The organization of behavior: A neuropsychological approach. John Wiley & Sons 1949
[5] Wills T. J. et al. Science 2005, 308(5723): 873-876.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-42-320.jpg)



![46
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
FRET‐based structural analysis of ion channel regulation at the nanoscale
René Ebrecht, Gertrude Bunt
Clinical Optical Microscopy, Institute of Neuropathology, University Medical Center
Göttingen
gbunt@gwdg.de
A precise and tight regulation of ion channel activity is a prerequisite for proper cellular
functioning. Regulatory mechanisms use the binding of several regulatory and modulatory
proteins to the channel, along with structural arrangements within the channel subunits.
Inhibition by Ca2+/CaM binding is one of the most important regulation mechanisms for the
voltage-dependent potassium channel eag [1]. The channel contains multiple intracellular
domains that mediate Ca2+-mediated calmodulin binding [2], but the structural mechanism
behind CaM-mediated channel inhibition is not yet fully understood. Here we show, using
FRET imaging for the binding of CaM to heag1, that the two C-terminal binding domains, BD-
C2 and BD-C1, are the predominant binding sites in the native channel. Both sites can bind
CaM independently. Deletion of the N-termini results in reduced CaM binding, however the
binding domain in the N-terminus is not involved. Here we show that the N- and C-termini of
the channel subunits, by their direct intermolecular interaction, cooperate in CaM Binding to
the C-terminal binding domains. A 'transverse' Interaction between the N- and C-terminal tails
of the channel subunits support the binding of calmoldulin to the binding sites at the C-
terminus, likely forming a structural pocket that is required for efficient binding.
(1) Schönherr et al. EMBO J. (2000), 19 (13):3263-71
(2) Ziechner et al. FEBS J. (2006), 273(5):1074-86](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-46-320.jpg)

![48
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Localization of cell adhesion points using dual color MIET
Anna Chizhik, Carina Wollnik, Daja Ruhlandt, Alexey Chizhik , Narain Karedla, Dirk Haehnel,
Ingo Gregor, Florian Rehfeldt, Jörg Enderlein
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
anna.chizhik@uni-goettingen.de
We present the result on axial localization measurements of cell adhesion points with nm
accuracy. We used the recently developed metal-induced energy transfer (MIET) imaging,
which allows us to measure the axial localization of a fluorophore with 2-3 nm accuracy [1].
The principle of MIET imaging is based on the energy transfer between a fluorescent molecule
and a metal surface, which results in the molecules de-excitation rate acceleration and can be
observed as a shortening of the molecule’s fluorescence lifetime [1,2]. Because energy
transfer rate is monotonically dependent on the distance of a molecule from the metal layer
within near first 200 nm, the fluorescence lifetime can be directly converted into a distance
between the emitter and metal surface within this range of distances. Here, for the first time
we present the results of the dual-color MIET measurements correlated with FRET imaging
[3]. This allows us to simultaneously measure the axial localization of actin filaments and
vinculin and to monitor the areas where the distance between actin and vinculin is within
FRET-range, that is does not exceed 10 nm. By combining the realms of MIET and FRET
microscopy we achieve unprecedented axial resolution based on absolute and relative values
obtained by these methods.
[1] Chizhik, A. I. et al. Nature Photon. 8, 124-127 (2014).
[2] Karedla N. et al. ChemPhysChem, 15, 705–711 (2014).
[3] Förster, Th. Ann. Physik 437, 55-75 (1948).](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-48-320.jpg)
![49
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Filling the usability gap: Bioinformatics solutions for Image-Scanning Microscopy,
Stochastic Optical Fluctuation Imaging, and Surface Single Molecule Experiments
Dirk Hähnel, Narain Karedla, Anna Chizhik, Alexey Chizhik, Simon Christoph Stein, Anja Huss,
Sebastian Isbaner, Qui Van, Ingo Gregor, Jörg Enderlein
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
dirk.haehnel@phys.uni-goettingen.de, www.joerg-enderlein.de
Recent years have seen a tremendous increase of new and novel methods in the field of
superresolution fluorescence microscopy. Furthermore even better methods for increasing
axial resolution of fluorescence imaging have been introduced by our group very recently. Our
group has developed powerful methods: Confocal Spinning Disc Image-Scanning Microscopy
(CSDISM)1,2, Superresolution Optical Fluctuation Imaging (SOFI)3,4,5,6, and Metal Induced
Energy Transfer (MIET)7,8. However, new microscopy techniques that provide not only
enhanced image quality and resolution, but they are also simple enough for finding broad
application. To bridge the ultimate usability gap for end-users, we present simple soft- and
hardware solutions for CSDISM and SOFI which enable potential users to implement them in
an easy and straightforward way into their existing microscopy systems. In the case of CSDISM,
we have integrated the method into the environment of the widely used and popular
MicroManager Open Source Imaging platform. This allows any researcher who already has a
commercial Confocal Spinning Disk microscope to easily implement the image-scanning
option and thus to double the spatial resolution. For SOFI, we have developed a dedicated
hardware based on a Freely Programmable Gate Array (FPGA) which converts, in real time,
image movies taken by high-speed CCD systems into SOFI cumulant images. Thus, all
algorithmic complexities and numerical workload of SOFI calculations are taken care of.
Furthermore we will present our recently developed software tool for smart automated single
molecule on surface experiments termed (SIMA). This is an effective tool to save time and
enables the researcher to conduct complex measurements. SIMA increases the comparability
of single molecule measurements, and reduces bleaching to the absolute possible minimum.
[1] Müller and Enderlein, “Image Scanning Microscopy”;
[2] Schulz, Pieper, and Clever, “Resolution Doubling in Fluorescence Microscopy with Confocal Spinning-Disk Image Scanning Microscopy”;
[3] Dertinger et al., “Achieving Increased Resolution and More Pixels with Superresolution Optical Fluctuation Imaging (SOFI)”;
[4] Dertinger et al., “SOFI-Based 3D Superresolution Sectioning with a Widefield Microscope”;
[5] Dertinger et al., “Advances in Superresolution Optical Fluctuation Imaging (SOFI).”; Dertinger et al., “Fluctuation Imaging ( SOFI )”
[6] Geissbuehler et.al., “Live-cell multiplane three-dimensional super-resolution optical fluctuation imaging”;
[7] Chizhik et.al. “Metal-induced energy transfer for live cell nanoscopy”;
[8] Karedla et.al. “Single-Molecule Metal-Induced Energy Transfer (smMIET): Resolving Nanometer Distances at the Single-Molecule Level”](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-49-320.jpg)
![50
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Long-term information storage by the collective dynamics of multi-synaptic
connections
Michael Fauth, Florentin Wörgötter, Christian Tetzlaff
1Bernstein Center for Computational Neuroscience, Göttingen
Excitatory synapses in cortex typically reside on dendritic spines. Although cortical synapses
play an important role in long-term memory, these spines undergo a remarkably high turnover
[1,2]. This poses the question how information can be stored on a variable substrate as
synapses. As a possible solution, we propose that information is stored and retained by the
collective dynamics of multiple synapses. Such a collective dynamics can already be found on
the connection between two neurons, which can consist of multiple synapses. More precisely,
the experimentally obtained distribution of the number of synapses on these connections,
which are bimodal with peaks at zero and multiple synapses, can only emerge from a collective
dynamics of the involved synapses [3]. Modelling studies showed that this collective dynamics
can emerge from the interaction of synaptic and structural plasticity [4,5] and that it can be
influenced by external stimulation such that the neurons become either unconnected or
connected with multiple synapses [5].
Here, we investigate the information storage and retention of these collective dynamics with
a simple stochastic model of structural plasticity, where synapses are created with a constant
probability and removed with a probability depending on the number of existing synapses and
the external stimulation. Using information theoretic measures, we show that the collective
dynamics yielding the bimodal distributions of the number of synapses enables information
retention on time scales orders of magnitudes longer than the typical lifetime of a synapse.
Thus, the conflict of spine turnover and long- term memory can be resolved by storing
information in the collective dynamics of multiple synapses. Yet, at different external
stimulation levels where the collective dynamics yield distributions with a single peak either
at zero or at multiple synapses, information about the initial conditions decays quickly. This,
however, implies that these stimulations can be used to learn new information orders of
magnitude faster than it is forgotten. We confirm this by using these stimulations to store an
image in a population of multi-synaptic connections. Indeed, this image can be retained orders
of magnitude longer than it took to store it. Thus, learning can be faster than forgetting, which
is also a necessary prerequisite to solve the plasticity-stability dilemma in learning and
memory on the time scale of structural changes.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-50-320.jpg)
![51
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
[1] Yang G, Pan F, Gan WB (2009) Stably maintained dendritic spines are associated with lifelong memories. Nature 462: 920-924.
[2] Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, et al. (2009) Rapid formation and selective stabilization of synapses for enduring motor memories.
Nature 462: 915-919
[3] Fares T, Stepanyants A (2009) Cooperative synapse formation in the neocortex. Proceedings of the National Academy of Sciences.
106:16463–16468.
[4] Deger M, Helias M, Rotter S, Diesmann M.(2012) Spike-timing dependence of structural plasticity explains cooperative synapse formation
in the neocortex. PLoS Comput Biol. 8:e1002689.
[5] Fauth M, Wörgötter F, Tetzlaff C (2015) The formation of multi-synaptic connections by the interaction of synaptic and structural plasticity
and their functional consequences, PLOS Comput Biol. 11(1):e1004031](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-51-320.jpg)

![53
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Functional and genetic dissection of mechanosensory organs of Drosophila
1, 2C. Guan, 1N. Scholz, 1R. J. Kittel, 1T. Langenhan
1Institute of Physiology – Neurophysiology, Julius-Maximilians-University Würzburg
2III. Institute of Physics – Biophysics, Georg-August-University Göttingen
chonglin.guan@phys.uni-goettingen.de
Larval chordotonal neurons provide fundamental sensory information as they convert
mechanical stimuli into biological responses (stretch, touch and sound). They are
monociliated, bipolar nerve cells that reveal genetic and functional parallels with inner hair
cells of the mammalian ear [1, 2]. Here we have developed a preparation to directly record
from sensory neurons of the lateral chordotonal organ (lch5) during mechanical stimulation.
This method enables to correlate the neuronal electrical output with defined mechanical
input. We have used this setup to characterize basal functional lch5 parameters including time
course of response during continuous mechanical stimulation and the recovery time between
successive bouts of stimulation.
Previously, we identified the calcium-independent receptor of α-latrotoxin
(dCIRL/Latrophilin), a member of the Adhesion class of G protein-coupled receptors (aGPCR),
as a mechanoreceptor [3]. We found that dCIRL modulates lch5 neuron activity by adjusting
the mechanogating properties of ionotropic receptors known to produce receptor potentials
that subsequently lead to the generation of nerve impulses. Furthermore, our results indicate
that the extent of the extracellular NTF of dCIRL shapes mechanosensitivity of the lch5. These
experiments provide new insights into the mechanobiology of dCIRL and establish
chordotonal organs as interesting sites to study the molecular machinery involved in the
perception of mechanical challenges.
[1] Eberl, D. F., Hardy, R. W. & Kernan, M. J. Genetically similar transduction mechanisms for touch and hearing in Drosophila. J Neurosci
20, 5981-5988 (2000)
[2] Nadrowski, B., Albert, J. T. & Gopfert, M. C. Transducer-based force generation explains active process in Drosophila hearing. Curr
Biol 18, 1365-1372, doi:10.1016/j.cub.2008.07.095 (2008)
[3] Scholz, N. et al. The Adhesion GPCR Latrophilin/CIRL Shapes Mechanosensation. Cell Rep 11, 866-874, doi:10.1016/j.celrep.2015.04.008
(2015)](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-53-320.jpg)
![54
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
The role of competition in memory organization
1Juliane Herpich, 1, 2Florentin Wörgöttier, 2Christian Tetzlaff
1
III. Institute of Physics – Biophysics, Georg August University Göttingen
2Bernstein Center for Computational Neuroscience, Göttingen
Humans are able to perform cognitive strategies to solve problems they are faced with. Thus,
they generate a huge variety of strategies which cannot all be hard wired in their neuronal
networks which consist of a finite number of neurons. One hypothesis is that neural entities
are reorganized to participate in different cognitive purposes. Therefore, different entities are
exploited, recycled, and redeployed and, thus, put to different uses without losing their
original function [1]. Given this idea, to enable an accurate reaction according to a given
situation, humans adaptively organize the learned memories of previous experienced
environmental stimuli. However, the neuronal principles for the functional reorganization of
the brain, thus, for rewiring the links between memories are still unknown.
Here, we use an adaptive neuronal network model depending on the interactions of synaptic
plasticity [2, 3] and synaptic scaling [4]. Hebbian synaptic plasticity adapts the efficacies of
synapses dependent on the corresponding neuronal activities [5]. With the intertwined
mechanism of synaptic scaling, thereby, synaptic plasticity yields the formation of strongly
interconnected subgroups of neurons (cell assemblies; CAs) [6]. These CAs serve as neuronal
representations or memories of specific environmental stimuli [5]. As we are interested in the
functional organization of the brain, we started to investigate the interaction between two
memories. We describe the dynamics for the representation of each memory by
homogeneous populations and drive the CAs with different external stimuli. It is shown that
neuronal competition (synaptic plasticity combined with synaptic scaling) is mandatory for the
formation of CAs [4, 7]. Here, we investigate the role of competition between memories for
their functional rewiring. Therefore, we combine synaptic plasticity with different generic
synaptic scaling mechanisms. Thus, we gradually increase the influence of synaptic scaling
from a constant to a more complex and activity-dependent condition. Interestingly, increased
competition between both CAs leads to the formation of different functional interactions
between them. Dependent on the external drive and the internal competition the two-
memory system is capable to build up different functional links between these memories such
as association, discrimination, and sequence [8].
This work describes different forms of functional organization of memories in the brain.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-54-320.jpg)


![57
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Single-molecule Metal-Induced Energy Transfer (smMIET): resolving nanometer
distances and dynamics at single molecule level
1,2Narain Karedla, 1Arindam Ghosh, 1Sebastian Isbaner, 1Roman Tsukanov, 1Alexey I. Chizhik,
1Ingo Gregor, 1,2Jörg Enderlein
1III. Institute of Physics – Biophysics, Georg August University Göttingen
2DFG Research Center Nanoscale Microscopy and Molecular Physiology of the Brain,
Göttingen
We present a new concept for measuring distance values of single molecules from a surface
with nanometer accuracy using the energy transfer from the excited molecule to surface
plasmons of a metal film [1]. We measure the fluorescence lifetime of individual dye molecules
deposited on a dielectric spacer as a function of a spacer thickness. By using our theoretical
model [2], the lifetime values are converted into the axial distance of individual molecules.
Similar to Förster resonance energy transfer (FRET), this allow emitters to be localized with
nanometer accuracy, but in contrast to FRET the distance range at which efficient energy
transfer takes place is an order of magnitude larger. Combining orientation measurements [3],
one can potentially employ smMIET to localize single emitters with a nanometer precision
isotropically, which will facilitate intra- and intermolecular distance measurements in
biomolecules and their complexes, circumventing the requirement of the knowledge of
mutual orientations between two dipole emitters which severely limits the quantification of
such distances from a conventional single-pair FRET (spFRET) experiment. Furthermore, due
to the distance dependent fluorescence quenching, one can use smMIET to measure the
dynamics of a polymer chain or an intrinsically disordered protein (IDP) up to submicrosecond
time scales (dynaMIET). Here we explore the potential of smMIET using designed DNA
structures like hairpins and holliday junctions and randomly labeled lipid bilayers.
[1] Karedla, N., Chizhik, A.I., Gregor, I., Chizhik, A.M., Schulz, O., Enderlein, J., ChemPhysChem, 15, 705-711 (2014).
[2] Enderlein J., Biophyical Journal, 78, 2151-8 (2000).
[3] Karedla, N., Stein, S. C., Hähnel, D., Gregor, I., Chizhik, A., & Enderlein, J., Physical Review Letters, 115, 173002 (2015).](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-57-320.jpg)
![58
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
DNA binding properties of the archaeal MCM complex studies using AFM
Amna Abdalla Mohammed Khalid
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
amna.abdalla-mohammed-khalid@phys.uni-goettingen.de
Understanding at the molecular level the mechanisms that govern DNA replication in
proliferating cells is fundamental to understand disease connected to genomic instabilities, as
genetic disease and cancer. A key step for DNA replication to take place, is unwinding the DNA
double helix and this carried out by proteins called helicases. We then are interested to study
helicase connected to replication process in eukaryotic: is MCM (mini chromosome
maintenance) complex, six homologous MCM proteins known as MCM2-7 [1], which form a
ring that is supposed to "load" onto the DNA using energy produced by ATP hydrolysis and
move across unwinding the double helix. In our study we usually use archaeal MCM from
Methanothermobacter thermautotrophicus as a model system [2].
Our main idea is to investigate the
conformational changes of the DNA deposited
on a mica surface upon the interaction with
MCM proteins complex by means of AFM
imaging in air and in liquid.
I will present the work done using AFM
imaging in air to understand the static
conformations of MCM-DNA interaction from
accurate analysis of AFM topographic images
and then in liquid to follow the interaction
dynamic.
MCM complex: Replication fork progression
[1] Costa A. and Onesti S. (2009) Mol. Biol. 44, 326-342.
[2] Miller, J. M. & Enemark, E. J. (2015) ARCHAEA.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-58-320.jpg)


![61
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Tracking of Transport Processes in Living Cells Using
Single-Walled Carbon Nanotubes
1Constantin Kohl, 2Sreenath Ravindran, 1Kengo Nishi, 2Shilpa Dilipkumer, 2Ravi Muddashetty,
2Akash Gulyani, 1Christoph F. Schmidt
1III. Institute of Physics – Biophysics, Georg August University Göttingen,
2Institute for Stem Cell Biology and Regenerative Medicine,
National Center for Biological Sciences, Bangalore
constantin.kohl@phys.uni-goettingen.de
In this study, a novel advantageous imaging method using infrared-fluorescent DNA-wrapped
single-walled carbon nanotubes (SWNT) is applied to target specific proteins and locations in
living cells. [1]
Semiconducting SWNTs are highly photostable, non-blinking and non-bleaching [2]. Hence,
using SWNTs for dynamic fluorescent tracking represents a promising approach to follow
specific dynamics in functioning cells. To observe the near-infrared fluorescence of SWNTs,
we have built a setup enabling the simultaneous use of visible and infrared wide-field
fluorescence microscopy, highspeed imaging and imaging of GFP/RFP tagged cells, in
conjunction with infrared spectroscopy [3]. We apply several methods to solubilize the
hydrophobic SWNTs in watery solutions and use biochemical linking methods to specifically
target SWNTs in the cells [1].We furthermore discuss procedures with which SWNTs can be
introduced into several cell types.
[1] Fakhri et al., Science 344, 1031-5 (2014)
[2] Boghossian et al., ChemSusChem 4, 848-863 (2011)
[3] Wessel, PhD thesis (2015)](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-61-320.jpg)

![63
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Super-resolution microscopy in planta and of plant proteins
Julian Lehmann, Rainer Hedrich, Markus Sauer, Dietmar Geiger
Julius-von-Sachs Institute for Biosciences, Molecular Plant Physiology and Biophysics,
Julius-Maximilians-University Würzburg
Julian.Lehmann@stud-mail.uni-wuerzburg.de, www.super-resolution.de
Super-resolution and high-resolution imaging methods like direct stochastic optical
reconstruction microscopy (dSTORM) [1] or structured illumination microscopy (SIM) [2] are
major tools to determine the distribution of proteins or investigate protein-protein
interactions. Currently, most of the research is done in mammalian cells or in animal tissue.
To establish super-resolution microscopy in planta, we use Arabidopsis thaliana (AT), a model
plant, to study the slow anion channel 1 (SLAC1), its homologs (SLAH1-SLAH4) and other plant
specific proteins. The SLAC/ SLAH family is known to be addressed by a multitude of stimuli,
including stress hormones [3, 4]. Under drought S-type anion channels in guard cells are
stimulated by abscisic acid (ABA) [5], which triggers a decrease in cell volume and turgor
pressure and thereby causing stomatal closure. Although qualitatively well described [6], the
knowledge about the spatio/temporal dynamics of anion channel activation via the ABA-
receptor complex remains elusive.
By the generation of various AT mutants, expressing different proteins with fluorescent
proteins or by immuno-staining, we could analyze the distribution of these proteins and
protein-protein interactions in different plant cells. SIM measurements of SLAH1 and SLAH3
show a colocalization in AT leafs, further FRET-FLIM measurements illustrate the physical
interaction of SLAH1 and SLAH3. These results confirm previous electrophysiological
measurements of these two anion channels [7]. Particularly SLAH2 is expressed in plant roots,
where we could show the distribution of SLAH2 especially in endodermal cells and in the
pericycle. SIM imaging of PIN [8] proteins show a polar distribution in the root tip and first
dSTORM measurements of immuno-stained microtubules could be established.
1. van de Linde, S., et al., Nature Protocols, 2011. 6(7): p. 991-1009.
2. Gustafsson, M.G.L. Journal of Microscopy-Oxford, 2000. 198: p. 82-87.
3. Roelfsema, M.R.G. and R. Hedrich New Phytologist, 2005. 167(3): p. 665-691.
4. Roelfsema, M.R.G., R. Hedrich, and D. Geiger Trends in Plant Science, 2012. 17(4): p. 221-229.
5. Levchenko, V., et al. Proceedings of the National Academy of Sciences of the United States of America, 2005. 102(11): p. 4203-4208.
6. Geiger, D., et al., Proceedings of the National Academy of Sciences of the United States of America, 2009. 106(50): p. 21425-21430.
7. Cubero-Font, P. et al. Curr Biol, 2016. 26(16): p. 2213-20.
8. Krecek, P., et al. Genome Biol, 2009. 10(12): p. 249.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-63-320.jpg)
![64
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Differential Geometry of Filaments and Polymers
Steffen Mühle, Jörg Enderlein
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
steffen.muehle@phys.uni-goettingen.de, www.joerg-enderlein.de
The motivation behind this project is to provide an analytical model concerning the dynamics
of individual freely-swimming peptides which has been measured in our group. In order to
achieve this, we make an alternative approach to full molecular dynamics simulations or
simple beads-models like the Rouse model, namely via differential geometry. The geometric
framework which was published by Goldstein et al. [1] is used as a basis for further studies.
The peptide is treated as an ideal elastic rod which can be both bent and twisted. Its dynamics
minimize a given elastic energy functional under the constraint of local inextensibility. Thus
the ideal rod relaxes towards an elastic reference state such as a straight, untwisted line. The
geometric equations are then closed by balancing the elastic force and torque densities with
linear viscous drag terms, implying zero Reynolds number in the overdamped regime. This
procedure reveals entirely intrinsic evolution equations for the rod's twist and bent densities
and quantifies the elastic interplay between them [2]. However, the described process has
been completely deterministic until now and hence the main goal of this project is to expand
the model and include thermal fluctuations. Other effects that we want to include in the
model are internal friction, hydrodynamic interactions, excluded volume effects and viscous
loads on one end of the peptide. The latter may also serve to model an experiment in which
the peptide is bound to a surface.
[1] R. E. GOLDSTEIN AND S. A. LANGER, Nonlinear Dynamics of Stiff Polymers, Physical Review Letters, 75 (1995), pp.1094–1097
[2] R. E. GOLDSTEIN, T. R. POWERS, AND C. H. WIGGINS, The Viscous Nonlinear Dynamics of Twist and Writhe, Physical Review Letters, 80
(1998), p. 9](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-64-320.jpg)
![65
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Influence of Input Timing Variance on the Performance of Reservoir Networks
1,2Timo Nachstedt, 1,2Florentin Wörgötter, 1,2Christian Tetzlaff
1 III. Institute of Physics – Biophysics, Georg August University Göttingen
2Bernstein Center for Computational Neuroscience, Göttingen
timo.nachstedt@phys.uni-goettingen.de
Reservoir networks are a well-established model of neural networks performing complex
time-resolved computations [1]. They are a model of the processes implementing working
memory in human and animals [2]. The working principles stem from the concept of non-
autonomous and non-linear transient systems [3]. While various implementations and
applications of reservoir networks have been shown, an understanding of their abilities and
limitations is missing. Typical tasks include additive or multiplicative noise in the input signals
or within the reservoir itself. In most tasks, the timing of the input signals is very precise or
even constant. In real-world situations, a network continuously interacts with other networks,
i.e. brain areas, or the environment. The signals received via these pathways do not
necessarily exhibit a reliable timing. Here, we investigate the consequences of abolishing
precise timing of input signals. We train the network by both the Echo State Approach [4] as
well as the FORCE-method [5]. In both cases the performance of the reservoir declines with
increasing input variance. The transient storage mechanism relies on small distances between
the trajectories evoked by stimuli. Noise in the input timing affects this storage mechanism.
In order to increase the distances between trajectories additional read-out signals maintaining
relevant memory content can be introduced. This way, the originally purely transient network
is turned into a system with multiple attractor states. We propose that optimal performance
is achieved if the maintenance of memory content and the production of complex output
trajectories is separately implemented by attractor states and transients, respectively.
[1] Lukosevicius M, Jaeger H. Comput. Sci. Rev. 2009, 3(3):127-149. doi: 10.1016/j.cosrev.2009.03.005
[2] Barak O, Tsodyks M. Curr. Opin. Neurobiol. 2014, 25:20-24. doi: 10.1016/j.conb.2013.10.008
[3] Carvalho A, Langa J, Robinson J. Discrete Continuous Dyn. Syst. Ser. B 2015, 20(3): 703-747. doi: 10.3934/dcdsb.2015.20.703
[4] Jaeger H. GMD Report No. 148, German National Research Center for Information Technology. 200.
[5] Sussillo D. Neuron 2009, 63(4):544-557. doi: 10.1016/j.neuron.2009.07.018](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-65-320.jpg)


![68
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Complementary lipopeptides CPE and CPK induce fusion of lipid membranes:
molecular mechanism of lipopeptide – membrane interaction
1Sarka Pokorna, 1Alena Koukalova, 1Radek Sachl, 2Nestor Lopez Mora, 2Aimee Boyle,
2Alexander Kros, 1Martin Hof
1J. Heyrovský Institute of Physical Chemistry, Academy of Sciences of the Czech Republic
Prague
2Leiden Institute of Chemistry, Leiden University, Leiden
Minimal model system, inspired by molecular recognition of native SNARE proteins, comprises
of complementary lipopeptide molecules CP12K4 and CP12E4. The two lipopeptides,
embedded in distinct lipid bilayers, interact with each other via coiled-coil of their E/K
peptides, bringing the two membranes in the close contact and inducing effective fusion in
vitro. [1,2] Designing of an efficient system, which might be useful for in vivo application, e.g.
drug delivery, requires a good understanding of molecular mechanism behind the fusion
event. Assuming the fusion is triggered by coiled coil interaction of two complementary
peptides E4 and K4, the efficiency of this process might be, among others, influenced by i)
(lipo)peptide – membrane interaction and ii) homoclustering of lipopeptides incorporated in
a membrane. These two phenomena were approached using FCS and FRET techniques,
revealing strikingly different behavior of the CP12E4 and CP12K4 within the membrane.
CP12K4 was shown to laterally compress the lipid bilayer and form aggregates in higher
concentration. Moreover, its peptide moiety has the tendency to interact with the lipid
headgroups significantly. None of that was observed for CP12E4. Further, mechanism of the
initial step of the fusion event was foreshadowed, i.e. binding of peptide K4 to vesicles
containing CP12E4.
[1] H. Robson Marsden, N.A. Elbers, P.H.H. Bomans, N.A.J.M. Sommerdijk, A. Kros, A reduced SNARE model for membrane fusion., Angew.
Chem. Int. Ed. Engl. 48 (2009) 2330–3. doi:10.1002/anie.200804493.
[2] F. Versluis, J. Voskuhl, B. van Kolck, H. Zope, M. Bremmer, T. Albregtse, et al., In situ modification of plain liposomes with lipidated coiled
coil forming peptides induces membrane fusion., J. Am. Chem. Soc. 135 (2013) 8057–62. doi:10.1021/ja4031227](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-68-320.jpg)



![72
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Modified Fluorescence Resonance Energy Transfer (FRET)
near metal-dielectric surfaces
12Benjamin Schreiber, 2Kareem Elsayad, 1Katrin G. Heinze
1Rudolf Virchow Center for Experimental Biomedicine, Julius-Maximilians-University,
Würzburg
2Advanced Microscopy Facility, Vienna Biocenter Core Facilities, Vienna
benjamin.schreiber@virchow.uni-wuerzburg.de
Fluorescence Resonance Energy Transfer (FRET) between two fluorescent probes is a powerful
technique to measure distances in biological systems. For donor-acceptor separations
distances below the FRET radius (typically less than 10nm) the energy transfer is efficient
enough to be detected, and with knowledge of some other geometrical parameters, it is
possible to calculate the distance between the so-called donor and acceptor molecule with a
very high accuracy. Beyond ~10nm the effect is generally too weak to be detected. For certain
research questions, however, longer “FRET distances” are desirable. It is well known that the
total emission and detected emission of emitters in the subwavelength range above metallic
surfaces and nanostructures are modified [1-3]. Several groups have been working on
exploiting these effects to enhance FRET distances and efficiencies [4-5].
Here we present preliminary data on how to amplify low FRET signals by using one particular
type of biocompatible metal and dielectric coated microscopy slides. The substrates are
designed to increase efficiency and detection range of FRET. One future key application of this
enhanced FRET technique will involve G-protein-coupled receptors (GPCRs) and their dynamic
functional behavior in membranes of cells cultured on our coated microscopy slides.
[1] Chance RR, Prock A, and Silbey R. "Molecular fluorescence and energy transfer near interfaces." Adv. Chem. Phys 37.1 (1978).
[2] Elsayad K, Urich A, Tan PS, Nemethova M, Small JV, Unterrainer K, Heinze KG, “Spectrally coded optical nanosectioning (SpecON) with
biocompatible metal-dielectric-coated substrates,” Proc Natl Acad Sci U S A 110, 20069-20074 (2013).
[3] Chizhik AI, Rother J, Gregor I, Janshoff A, Enderlein J, “Metal-induced energy transfer for live cell nanoscopy,” Nat Photonics 8, 124-127
(2014).
[4] Ghenuche P, de Torres J, Moparthi SB, Grigoriev V, Wenger J, “Nanophotonic Enhancement of the Forster Resonance Energy-Transfer
Rate with Single Nanoapertures,” Nano Lett 14, 4707-4714 (2014).
[5] Yu YC, Liu JM, Jin CJ, Wang XH, et al. "Plasmon-mediated resonance energy transfer by metallic nanorods." Nanoscale Research Letters
8:209 (2013).](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-72-320.jpg)

![74
BBTS 2016 - BIOPHYSICS BY THE SEA 2016
Fluorescence Spectroscopy, Microscopy and Molecular Cell Mechanics and Theoretical Neurophysics
Study Conformational Dynamics of Intrinsically Disordered Protein by PET-FCS
Man Zhou, Qui Van, Ingo Gregor, Jörg Enderlein
III. Institute of Physics – Biophysics, Georg-August-University Göttingen
man.zhou@phys.uni-goettingen.de, www.joerg-enderlein.de
Intrinsically disordered proteins (IDPs) are proteins which lack a well-defined three-
dimensional structure. The abundance and functional significance of IDPs has been recognized
only recently [1]. Due to their properties, IDPs play an important role in cellular functions.
They serve as flexible inter-protein linkers, and participate in molecular recognition, molecular
assembly, cellular signaling and regulation, or protein modification. Thus, genetically encoded
alterations of IDPs are involved in many diseases, such as cancer, cardiovascular disease,
amyloidosis, or neurodegeneration [2]. Therefore the study and characterization of the
conformational dynamics of IDPs are important to better understand the underlying
mechanisms which lead to various pathologies.
FG repeats, rich in phenylalanine (F) and glycine (G), are one particular type of IDPs. FG repeats
are located in the central channel of the nuclear pore complex (NPC), and they control the
molecular transport between the nucleus and the cytoplasm [3]. The way how FG repeats
form or/and function as highly selective barriers in NPCs is not clear. Here, the conformational
dynamics of the FG repeat Nsp1 is investigated by photo-induced electron-transfer
fluorescence correlation spectroscopy (PET-FCS) and molecular dynamics simulation (MD).
Combination of PET-FCS and MD simulation offers a more comprehensive understanding of
the relationship between functional mechanism and conformational dynamics of IDPs.
The results from PET-FCS measurements indicate that the N-terminus of Nsp1 tends to be
more flexible than the C-terminus. Furthermore, short Nsp1 fragments (up to 50 amino acids)
at low concentration (100 μM) do not tend to aggregate under physiological condition. These
data indicate that the interaction between short FG repeats is not strong enough to solely
generate the barrier. The data of MD simulation showed that the conformations obtained by
the force field CHARMM 22* and a charm-modified TIP3P water model agrees best with the
experimental data. These results are important for further force field developments of MD
simulation for IDPs in the future.
[1] P. Tompa, “Intrinsically disordered proteins: a 10-year recap.,” Trends Biochem. Sci., vol. 37, no. 12, pp. 509–16, Dec. 2012.
[2] P. E. Wright et al., “Intrinsically disordered proteins in cellular signalling and regulation,” Nat. Rev. Mol. Cell Biol., vol. 16, no. 1,
pp. 18–29, Dec. 2015.
[3] F. Alber et al., “The molecular architecture of the nuclear pore complex.,” Nature, vol. 450, no. 7170, pp. 695–701, Nov. 2007.](https://image.slidesharecdn.com/biophysicsbythesea2016finalprogrambookprint-161217104608/85/Biophysics-by-the-sea-2016-program-and-abstract-book-74-320.jpg)