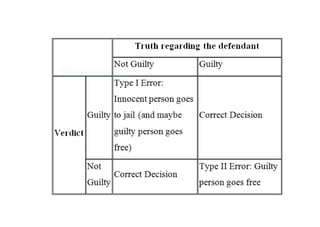
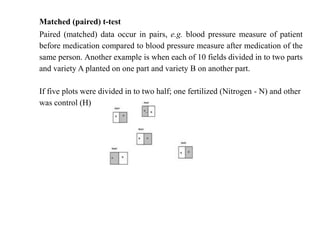
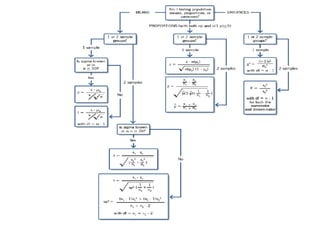
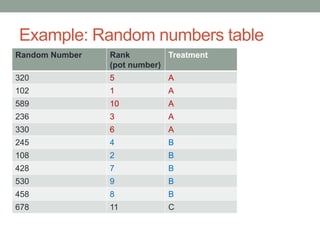
This document defines biometry and summarizes statistical methods for estimating population parameters from sample data. It begins by defining biometry as the application of statistical methods to biological problems, involving the measurement of life. It then discusses two types of estimation: point estimation, which provides a single value as an estimate, and interval estimation, which provides a range of values that the parameter is expected to fall within at a given confidence level. The document provides formulas and examples for constructing confidence intervals to estimate a single mean, the difference between two population means, and other parameters, depending on whether the population standard deviation is known or estimated from sample data, and whether sample sizes are large or small.
















































































































































































![Main effects are the averages of the simple
effects
• A main effect is the effect of an independent variable on a
dependent variable averaging across the levels of any other
independent variables.
• It is the average of the simple effects of a given independent
variable
From the above example,
• Main effect of variety = ½ (Simple effect of A at bo + Simple
effect of A at b1)
= ½ [(a1b0-a0b0) + (a1b1-a0b1)] = ½ [(2-1) + (4-1)] = 2
• Main effect of nitrogen = ½ (Simple effect of factor B at ao +
Simple effect of factor B at a1) = ½ [(a0b1-a0b0) + (a1b1-a1b0)] =
½ [(1-1) + (4-2)] = 1](https://image.slidesharecdn.com/biometryc1-230927132937-b7b59527/85/BIOMETRYc-1-pptx-182-320.jpg)

![Interaction
• Interaction is calculated as the average of the difference
between the simple effects.
• From the above example, it is calculated as the average of
the difference between simple effects of A at the two levels of
B or the difference between the simple effects of B at the two
levels of A.
= ½ (Simple effect of A at b1 – simple effect of A at b0)
= ½ [(a1b1-a0b1) - (a1b0-a0b0)] = ½ [(4-1) - (2-1)] = 1
OR
= ½ (Simple effect of B at a1 – simple effect of B at a0)
= ½ (a1b1-a1b0) - (a0b1-a0b0)] = ½ [(4-2) - (1-1)] = 1](https://image.slidesharecdn.com/biometryc1-230927132937-b7b59527/85/BIOMETRYc-1-pptx-184-320.jpg)









































































































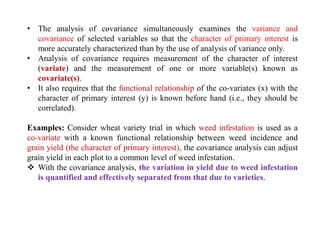









![Compute the relative efficiency (R.E.) of covariance analysis compared to standard analysis of
variance
R.E. = 100
)
1
)(
(
)
(
X
of
SS
Error
X
of
MS
Treat
Y
of
MS
adjusted
Error
Y
of
MS
unadjusted
Error
100
)
86.2
972.7/9
1
(
)
51.4
(
1608.7/18)
(
100
115.84
37
.
89
= 77.15%
Thus, the result indicates that the use of % dry matter as the covariate has
not increased precision in ascorbic acid content which would have been
obtained had the ANOVA is done without covariance.
CV = 100
x
Y
of
Mean
Grand
Y
of
MS
Error
Adjusted
= 100
94.63
51.4
= 7.6%
Mean comparison
S
d to compare two adjusted treatment means:
]
)
(
2
[
2
X
of
SS
Error
xj
xi
r
Y
of
Square
Mean
Error
Adjusted
, where xi & xj are the covariate means of ith
and jth
treatment; r is the number of replications common to both treatments.
For instance, to compare means of T1 & T2:
S
d = ]
86.2
)
34
-
43.67
(
3
2
[
51.4
2
where 34 & 43.67 are the covariate means of 1st
and 2nd
treatments;
3 is the number of replications common to both treatments.
S
d = 02
.
90 = 9.49](https://image.slidesharecdn.com/biometryc1-230927132937-b7b59527/85/BIOMETRYc-1-pptx-300-320.jpg)





![12.2 Simple Linear Regression and Correlation Analysis
12.2.1 Simple linear regression analysis
The simple linear regression analysis deals with the estimation and test of significance
concerning the two parameter α and β in the equation:
Y = α + βx
The data required for the application of the simple linear regression are the n-pairs (with
n >2) of y and x values.
Steps to estimate α and β
Step 1: Compute the means (x and y ), deviation from means [
x
x ,
y
y ], square of the
deviates (x2
, y2
) and product of deviates (xy).](https://image.slidesharecdn.com/biometryc1-230927132937-b7b59527/85/BIOMETRYc-1-pptx-306-320.jpg)












