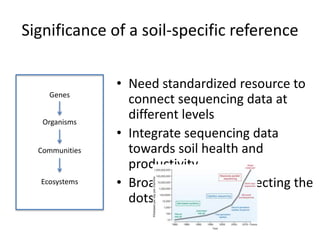
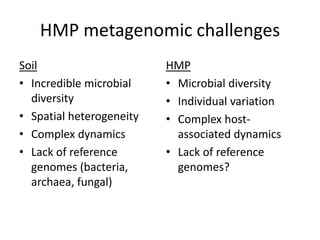
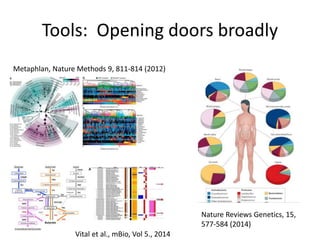
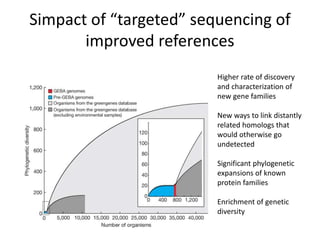
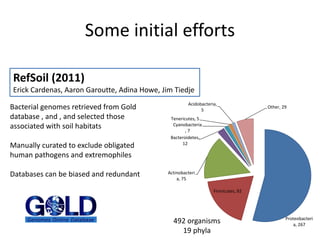
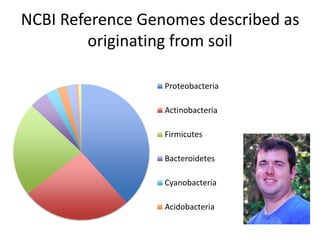
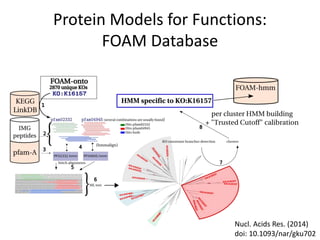
This document discusses the need for a soil-specific reference database to connect soil metagenomic sequencing data at different levels and enable broader understanding of soil health and productivity. It outlines challenges in soil metagenomics like incredible microbial diversity and lack of reference genomes. Lessons are drawn from the Human Microbiome Project reference genome effort. A targeted sequencing approach could benefit soil studies by providing frameworks for sequencing and identification. Challenges include defining soil organisms and important metadata. Initial efforts like RefSoil and NCBI Reference Genomes from soil are noted but require further curation. Contributions to developing such a database are welcomed.