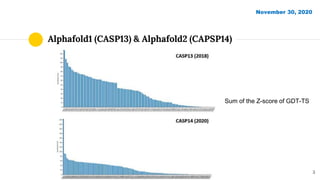
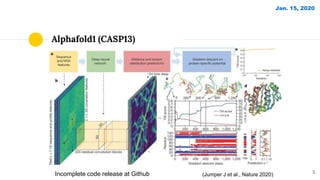
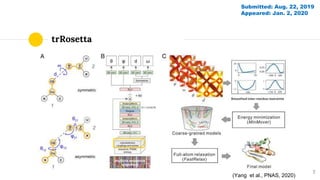
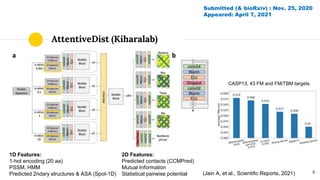
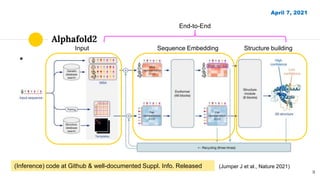
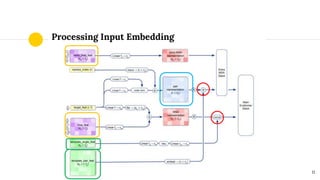
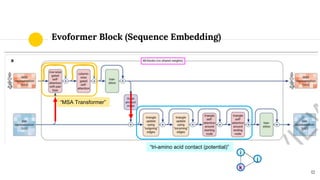
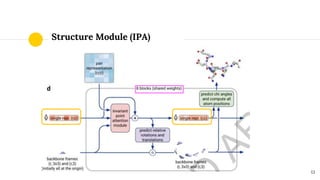
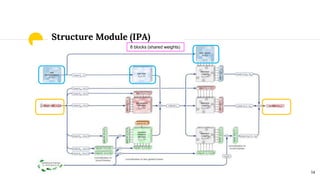
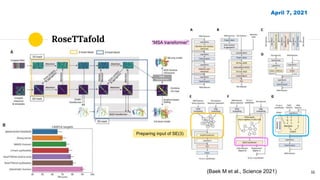
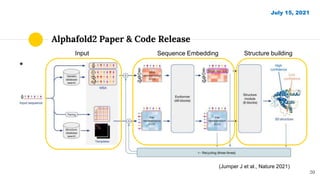
The document discusses advancements in protein structural bioinformatics with a focus on AlphaFold2, highlighting its development after AlphaFold1 and its significant contributions to protein structure prediction. Key methodologies, such as sequence embedding and multi-sequence alignment processing, are detailed along with the model's applications in areas like protein docking and drug screening. Future directions for research following AlphaFold2 are also suggested, emphasizing ongoing developments in structural modeling and computational techniques.