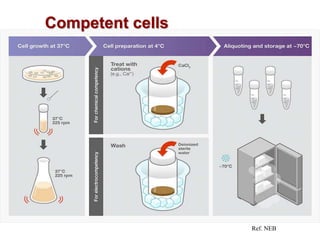
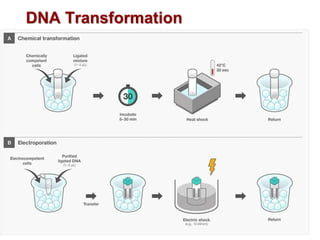
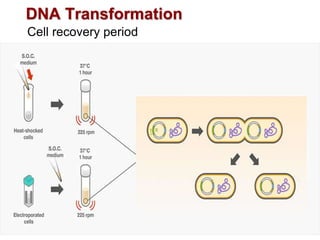
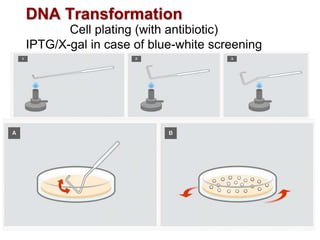
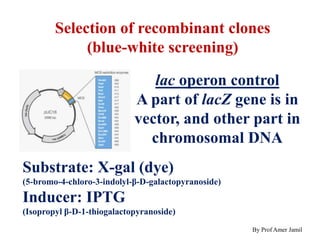
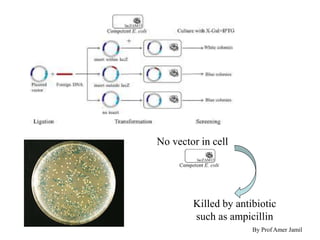
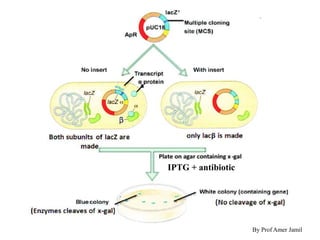
This document describes the process of constructing a recombinant DNA molecule:
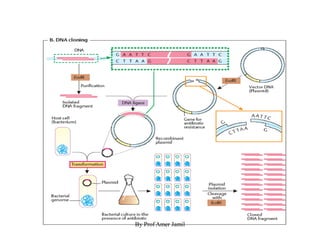
1. DNA/RNA is isolated and the gene of interest is amplified via PCR or RT-PCR.
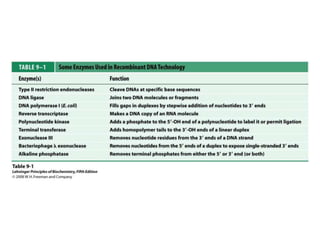
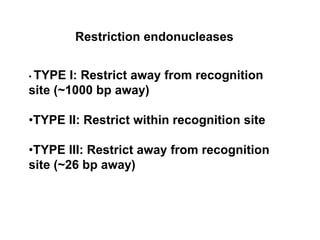
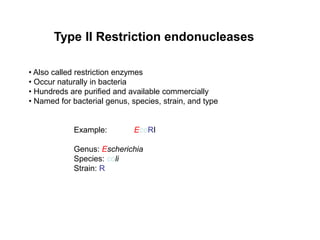
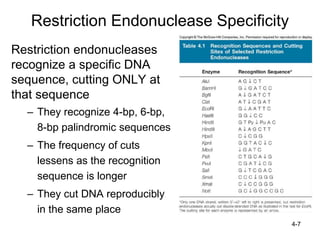
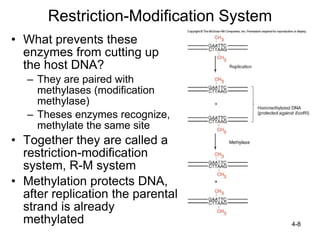
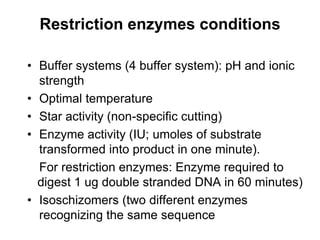
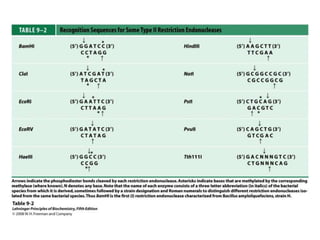
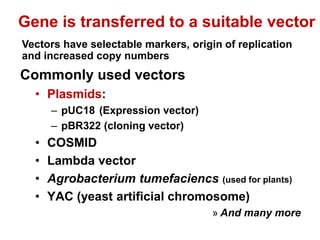
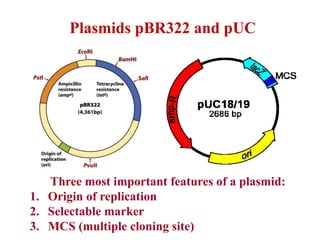
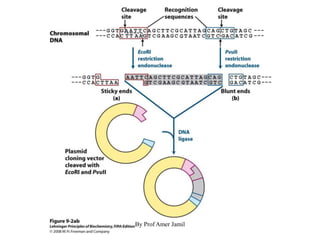
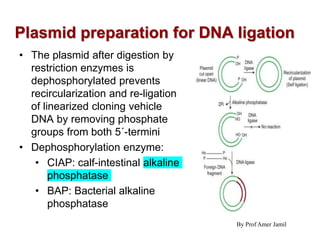
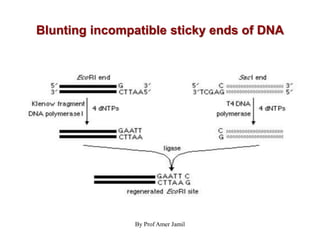
2. Restriction enzymes are used to cut the DNA molecule into fragments that are inserted into a plasmid vector.
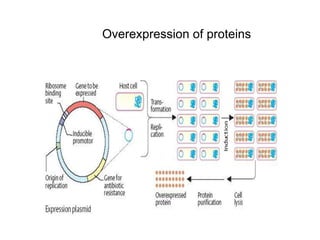
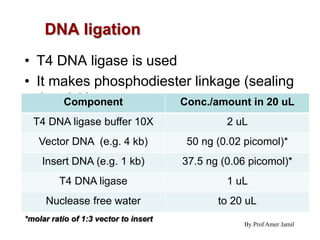
3. The recombinant molecule is transformed into E. coli host cells which then produce many copies of the molecule that can be isolated.