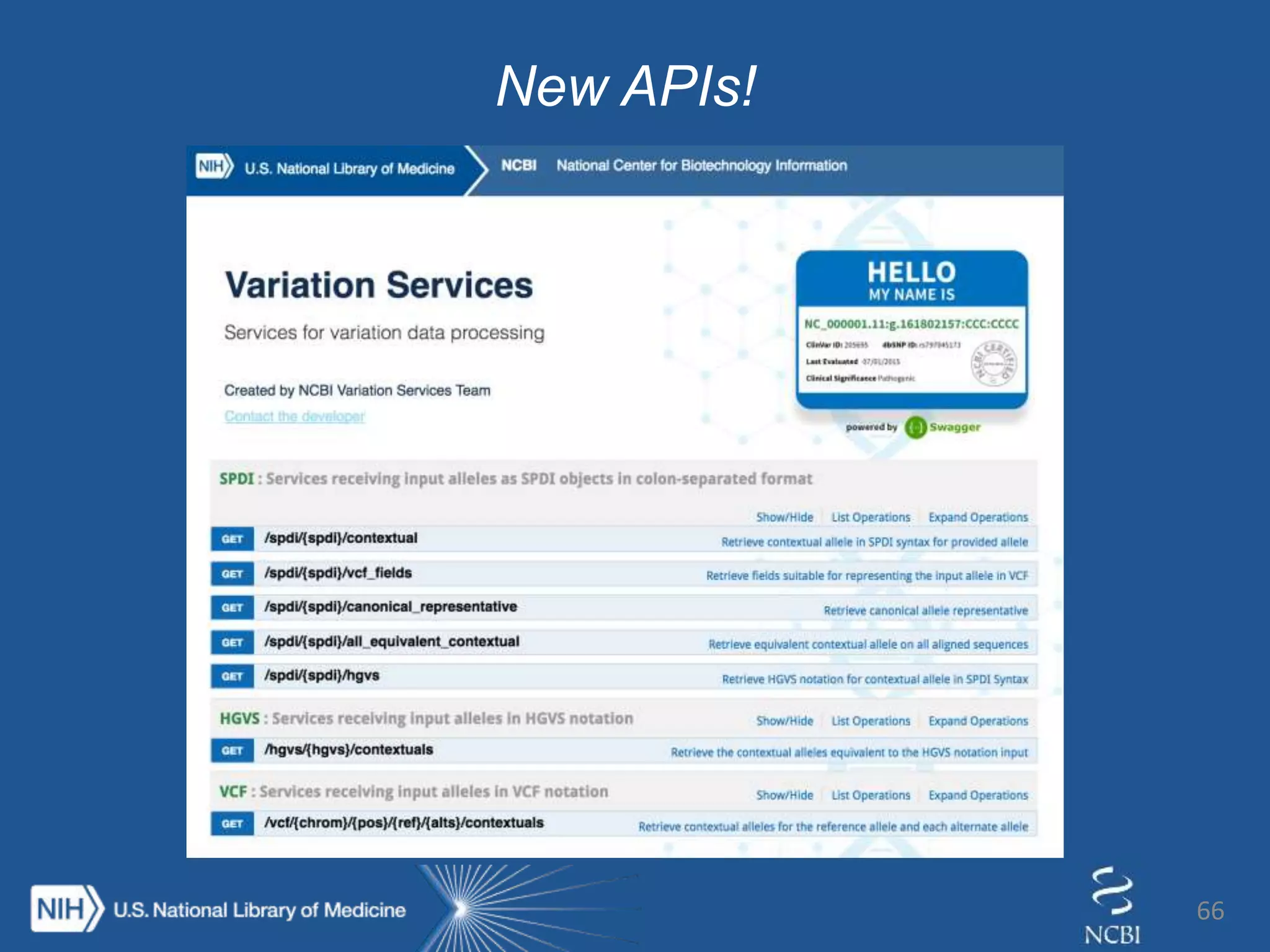
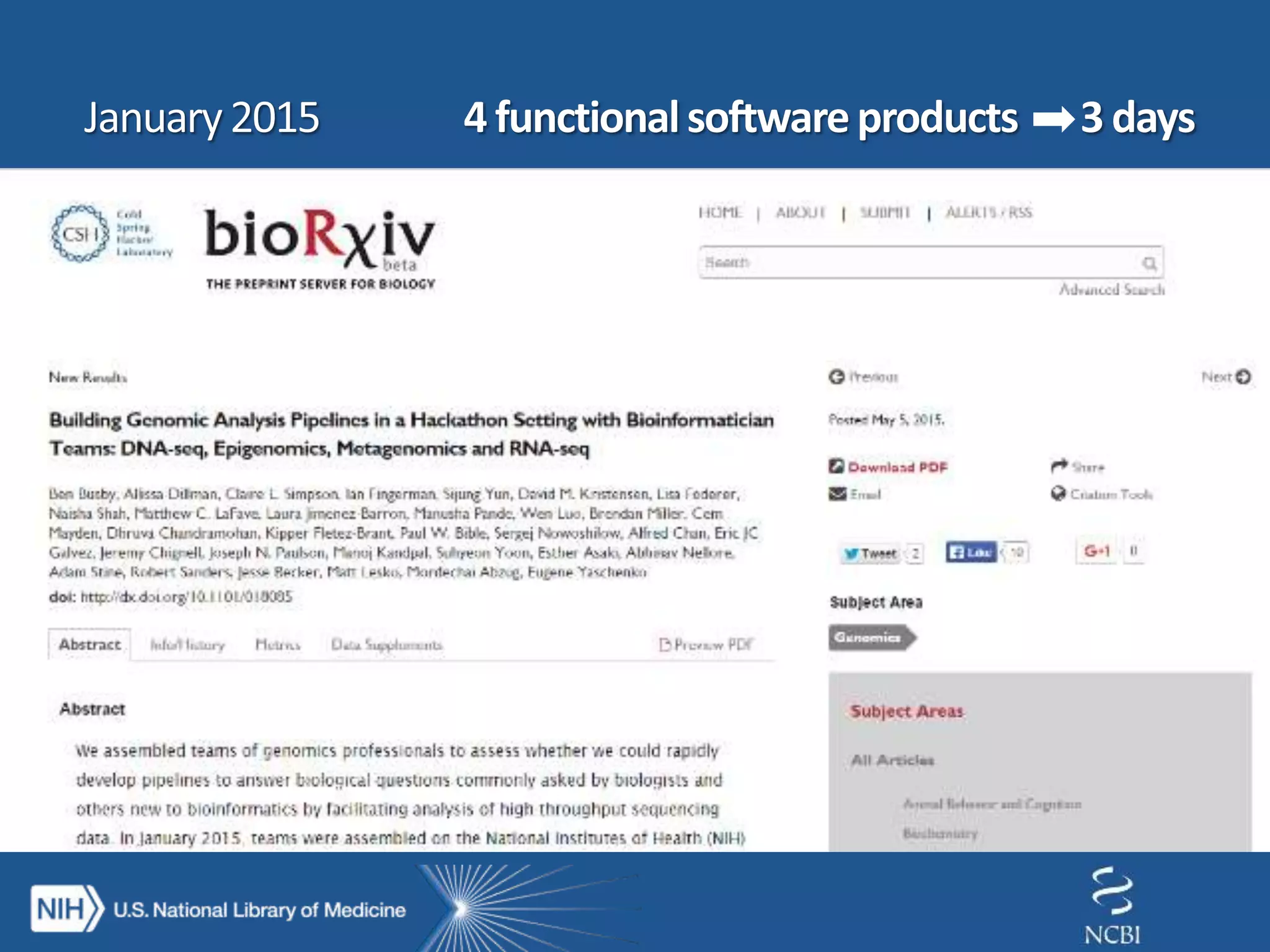
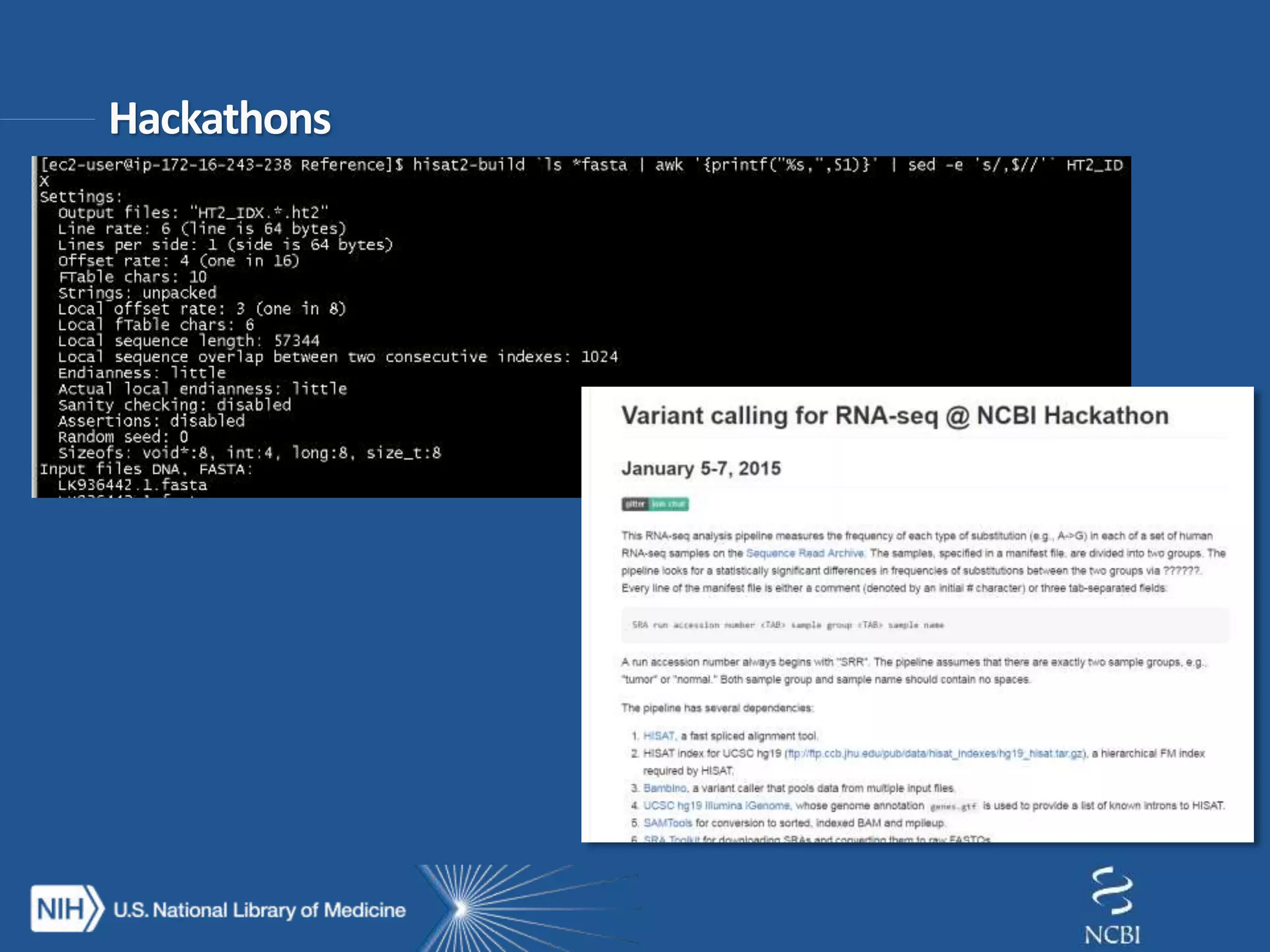
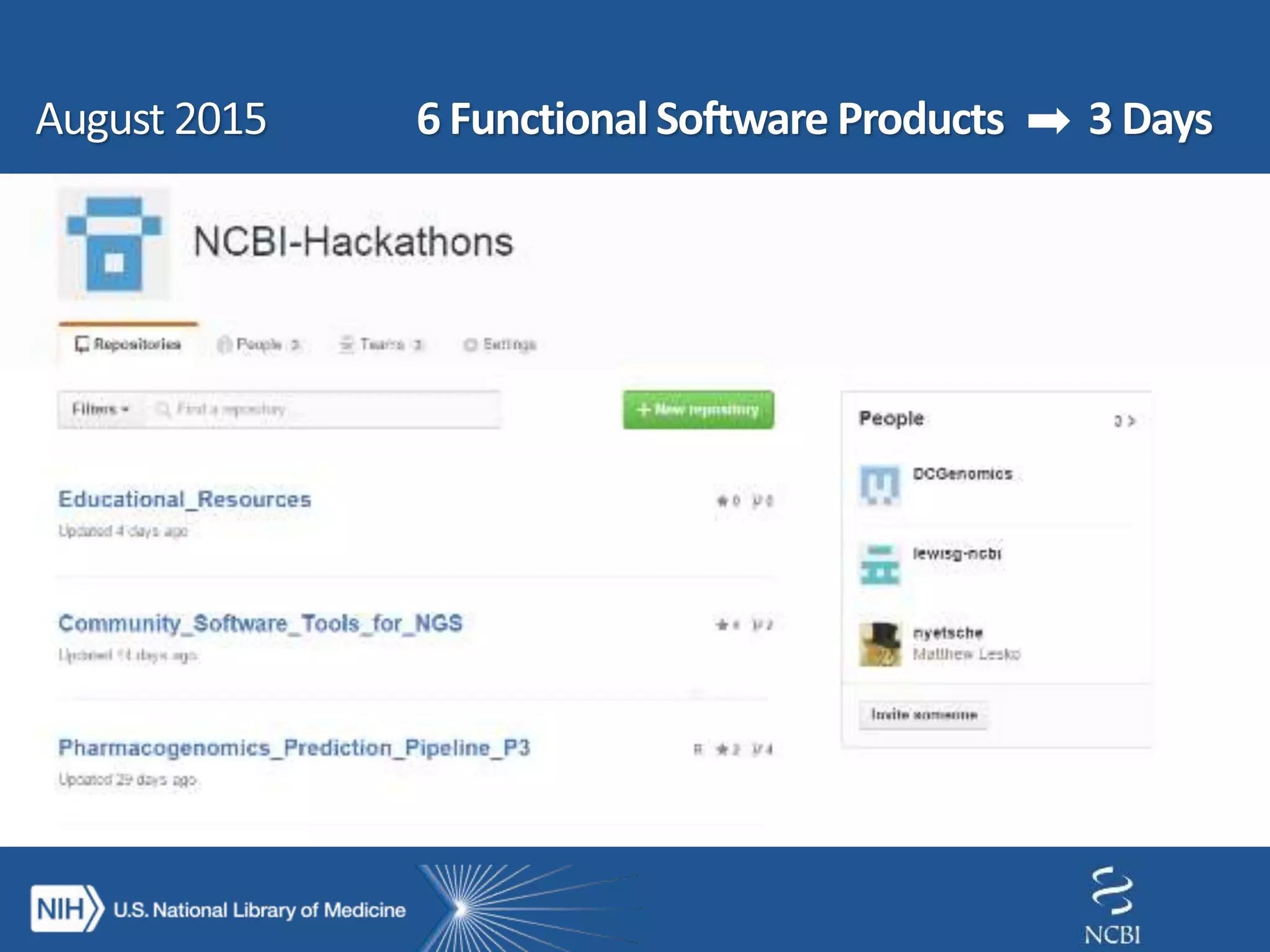
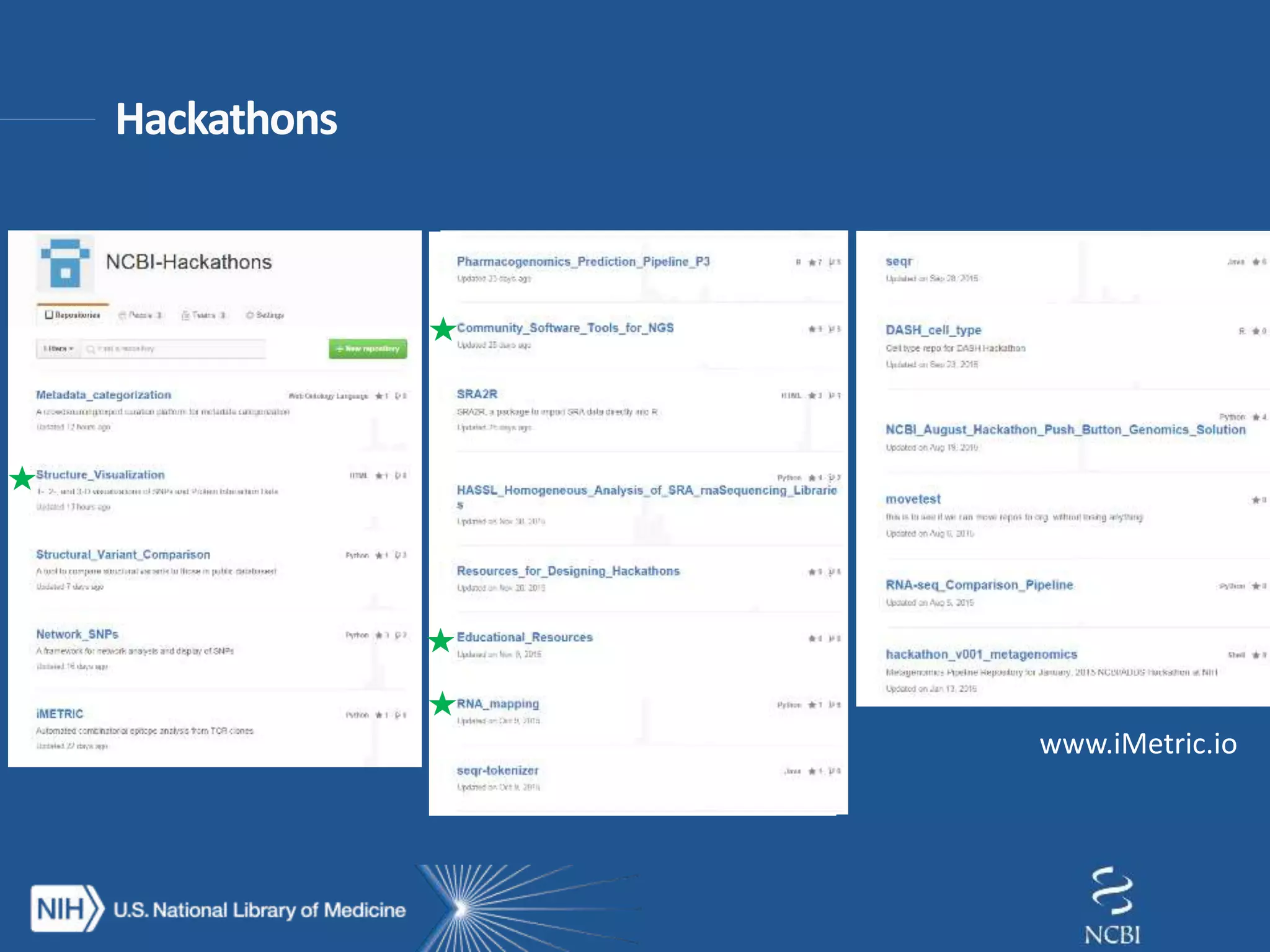
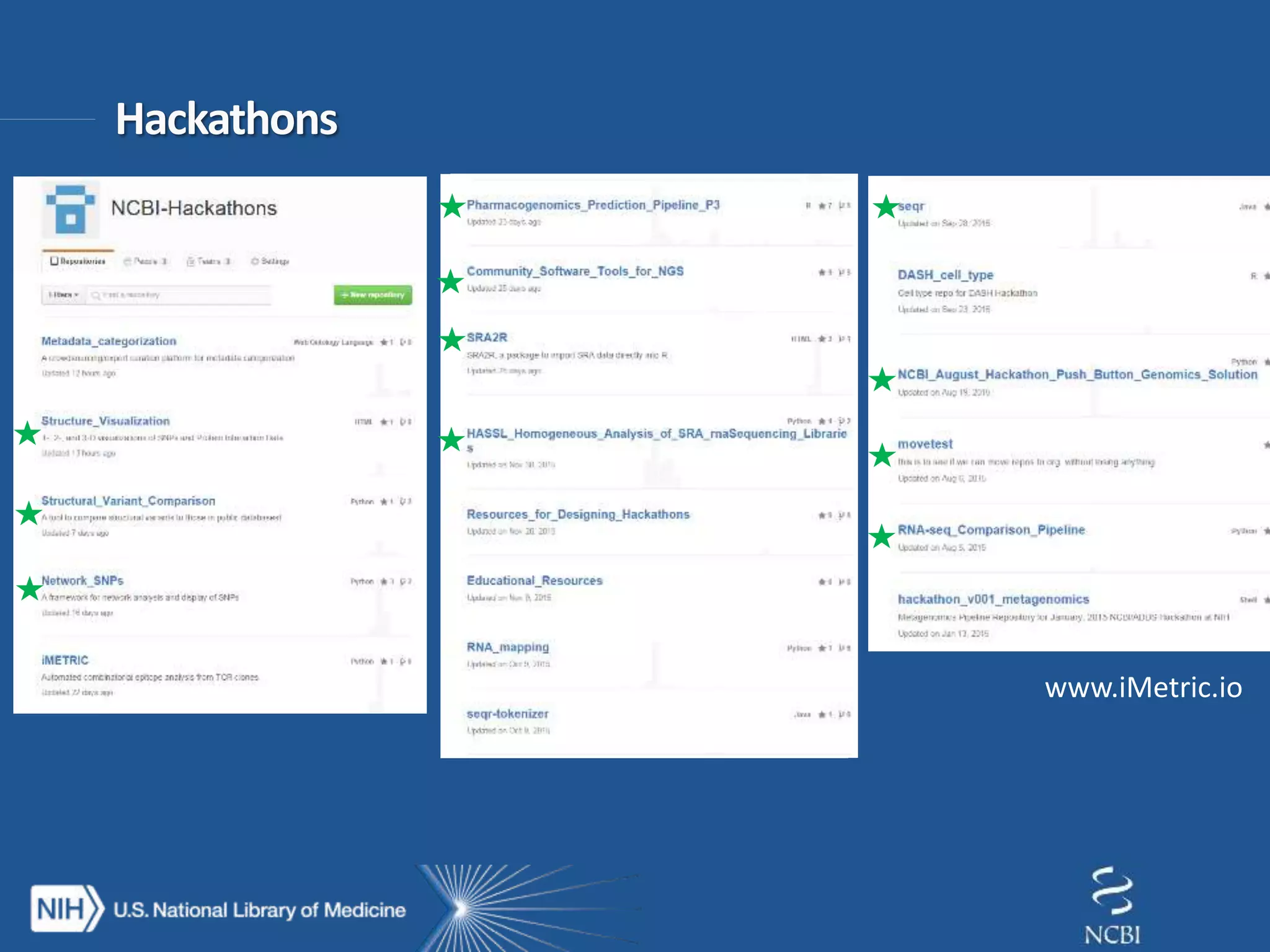
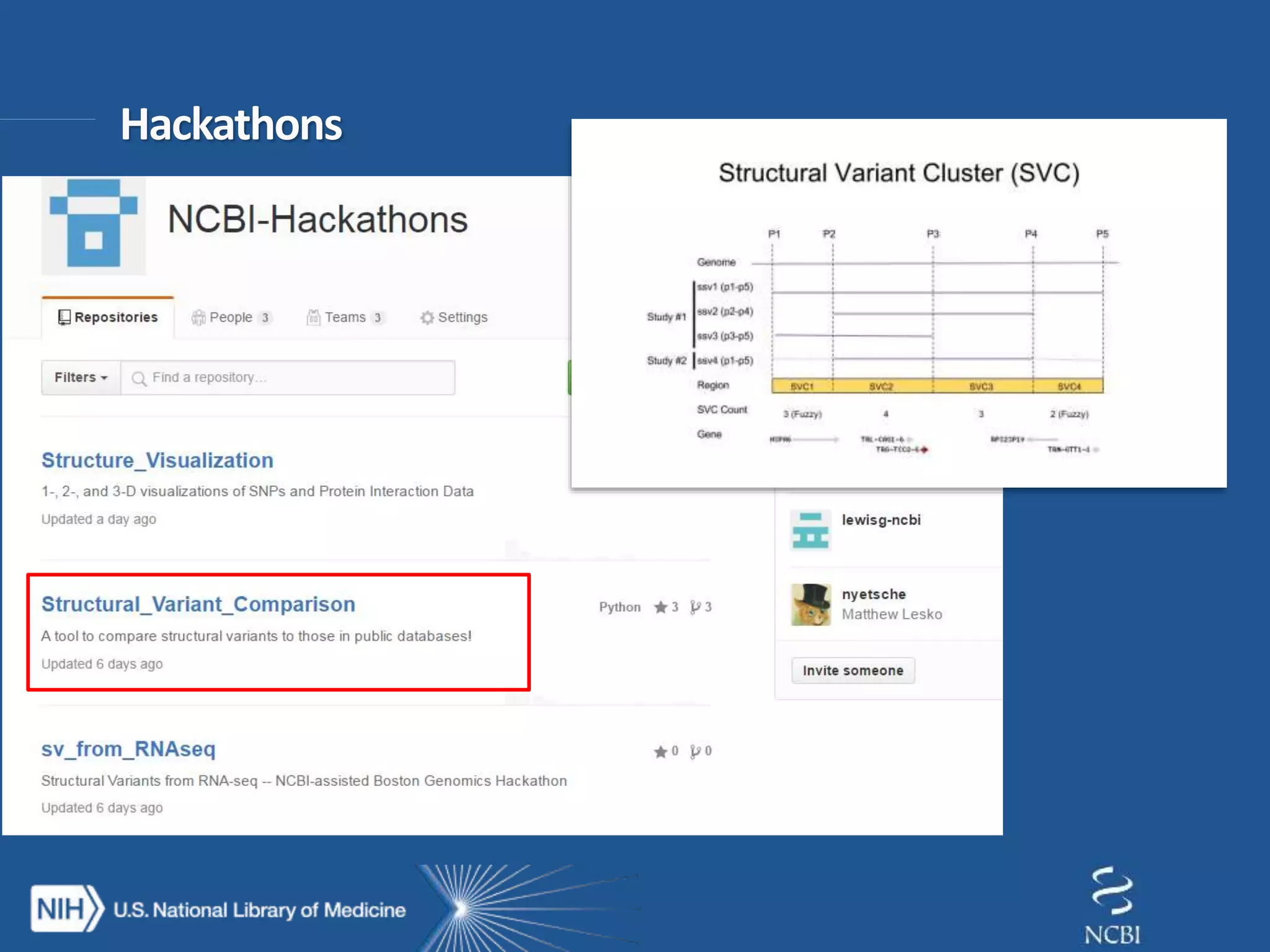
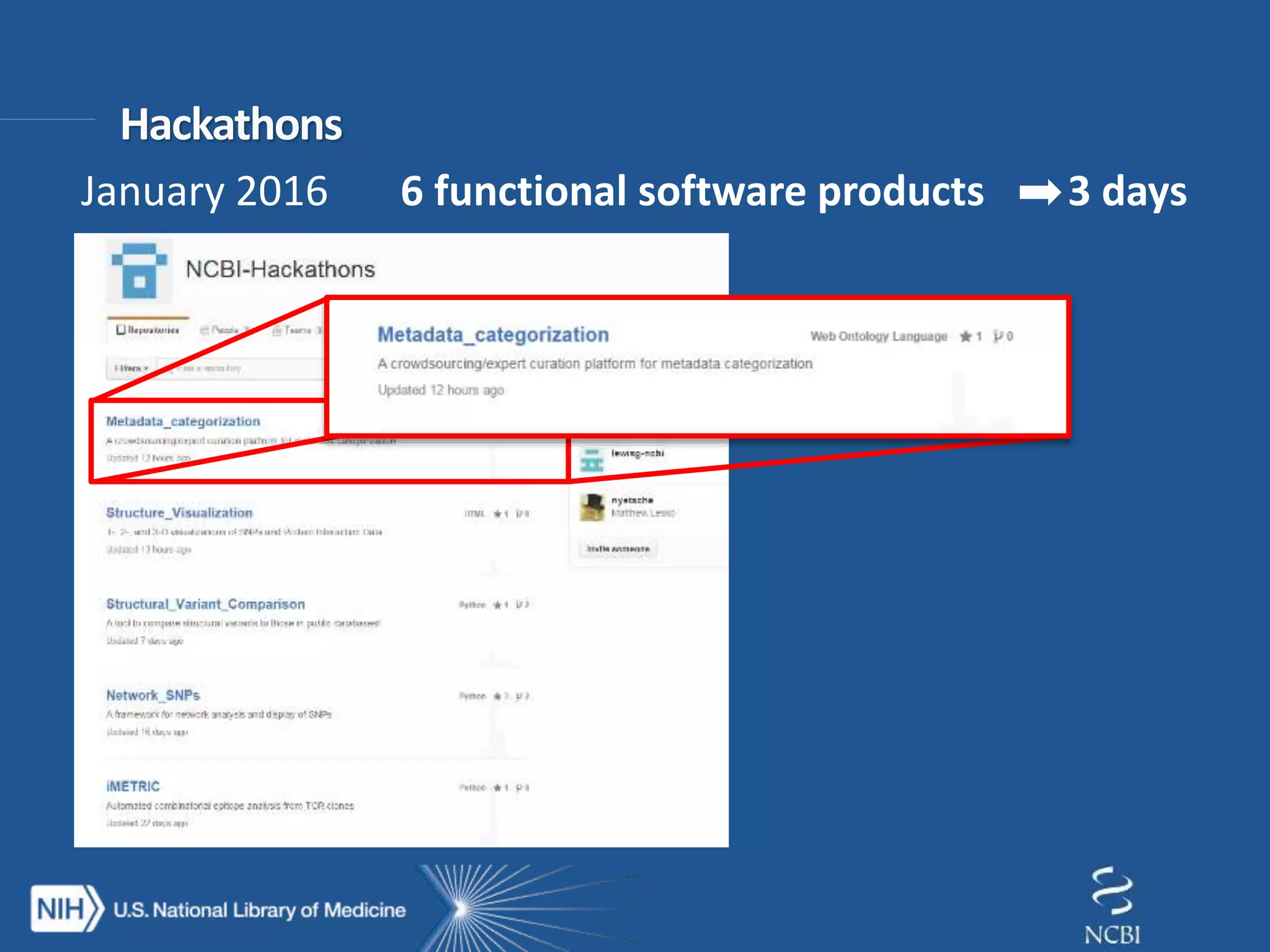
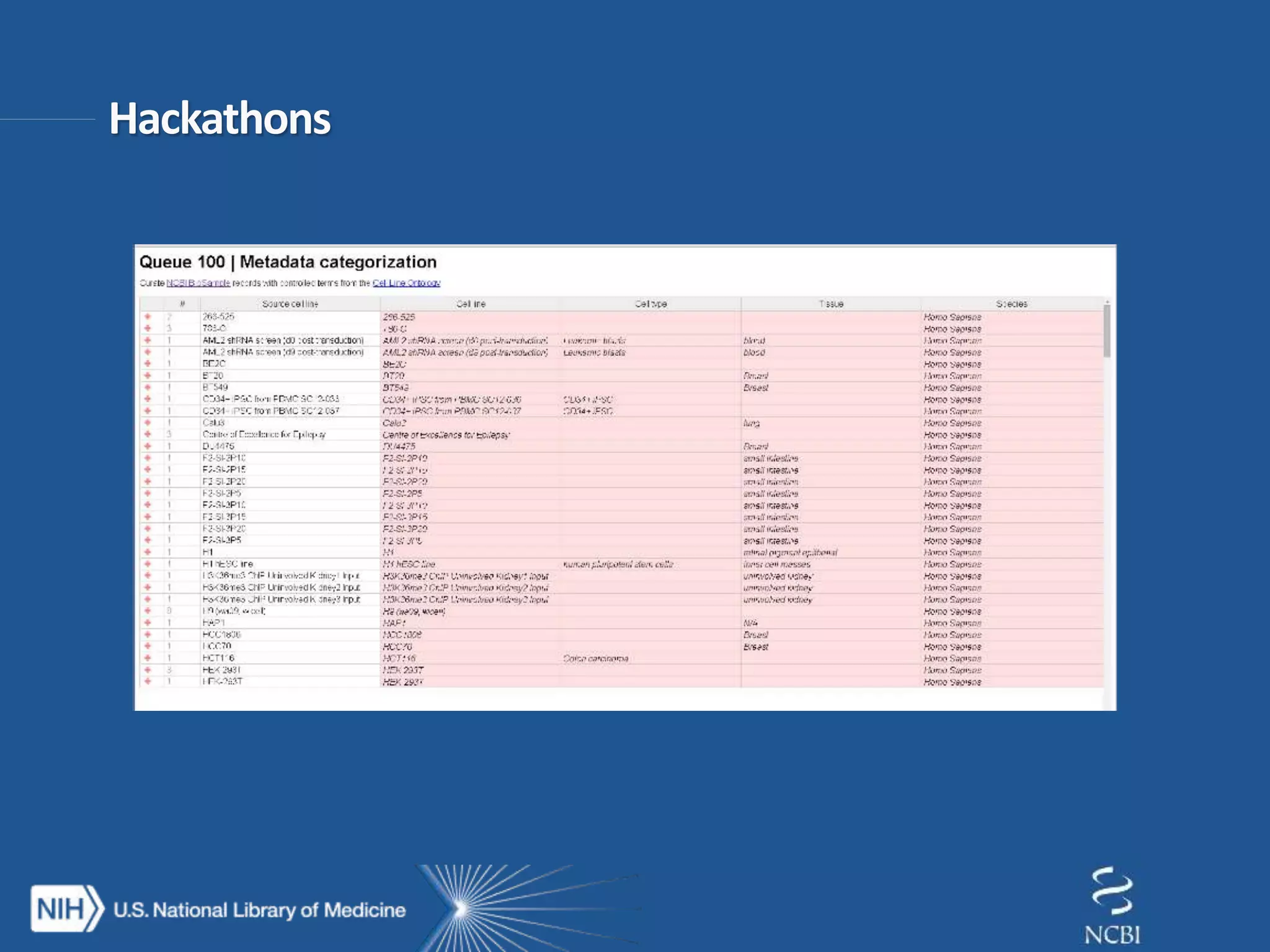
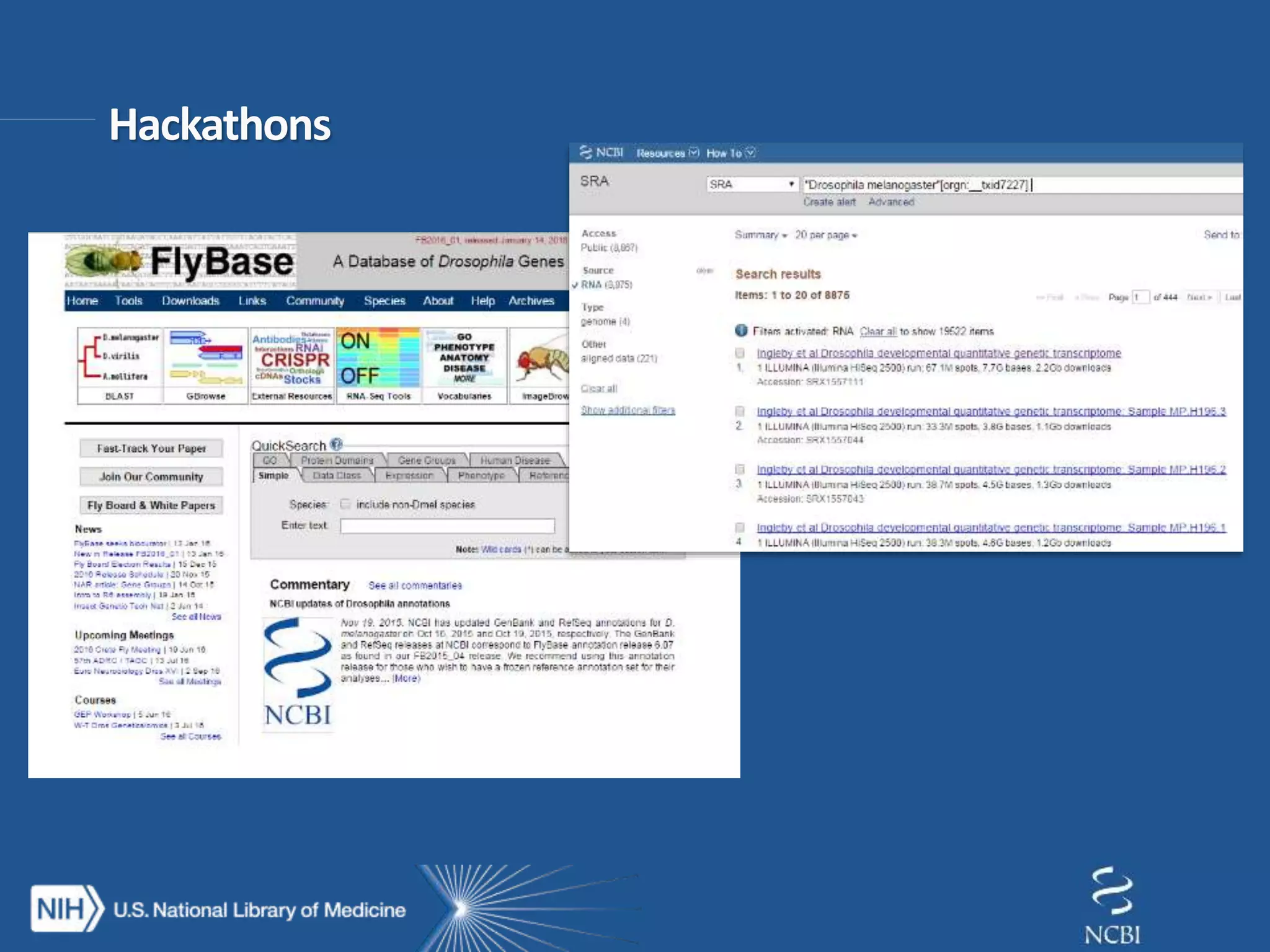
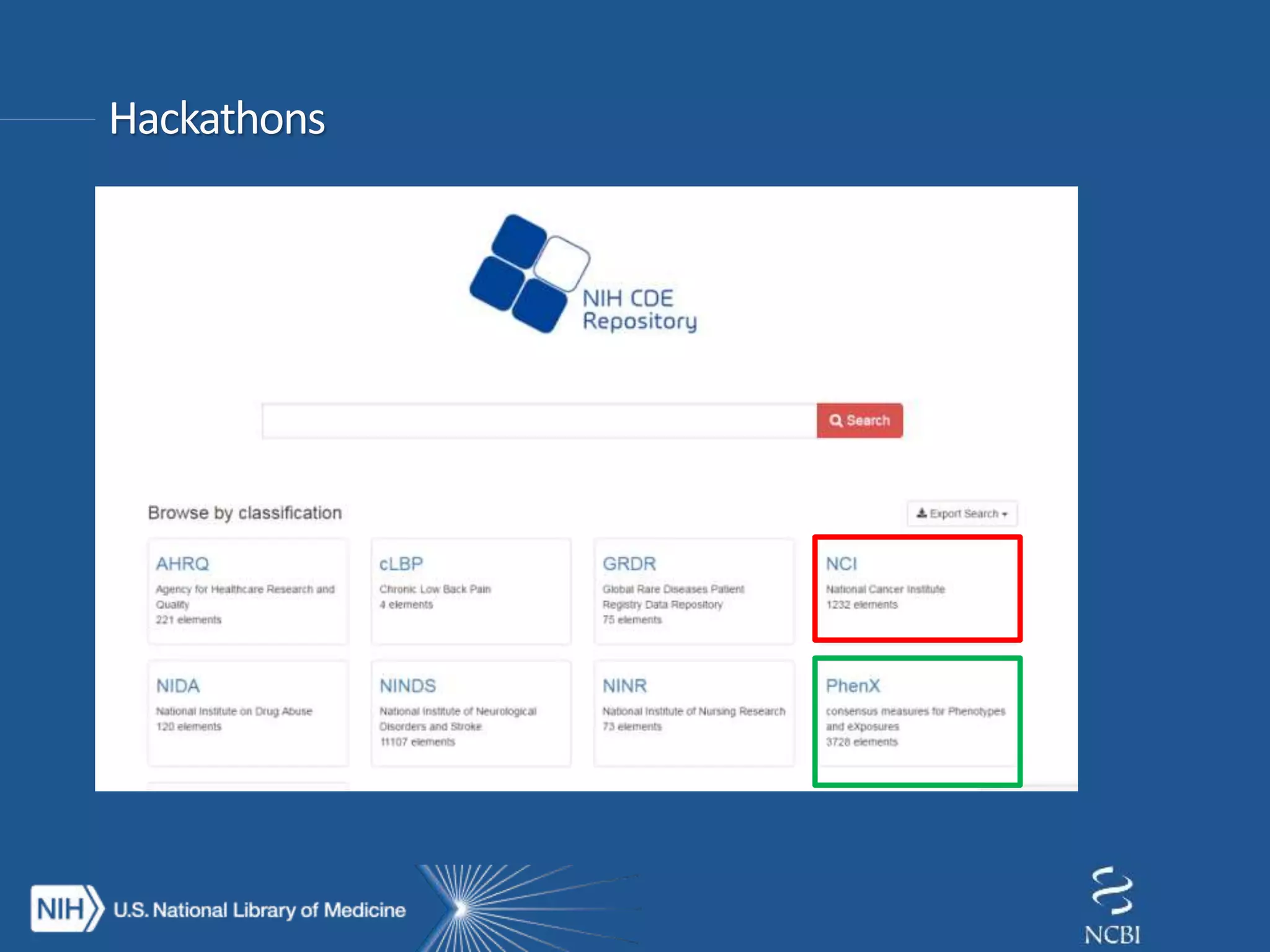
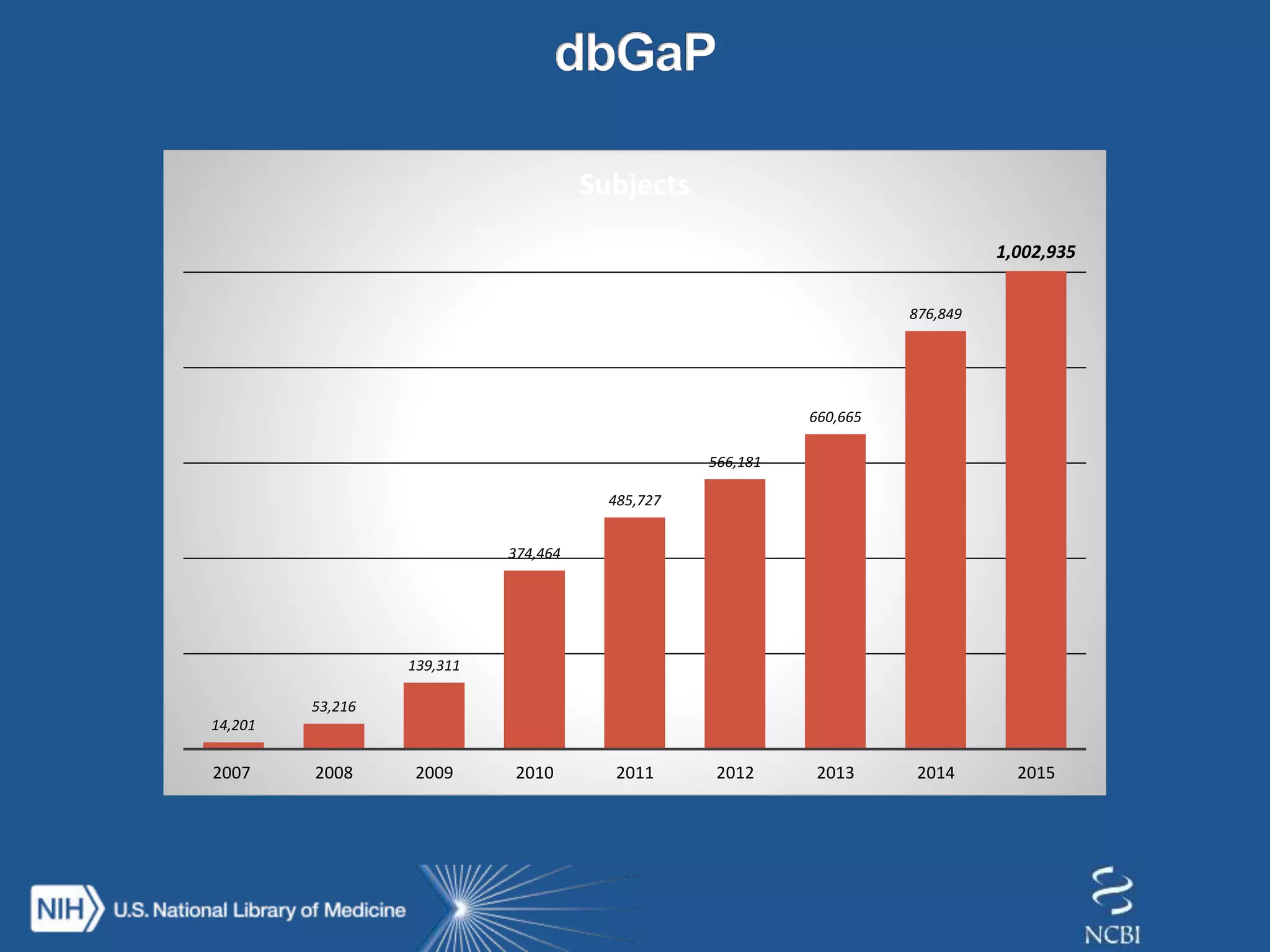
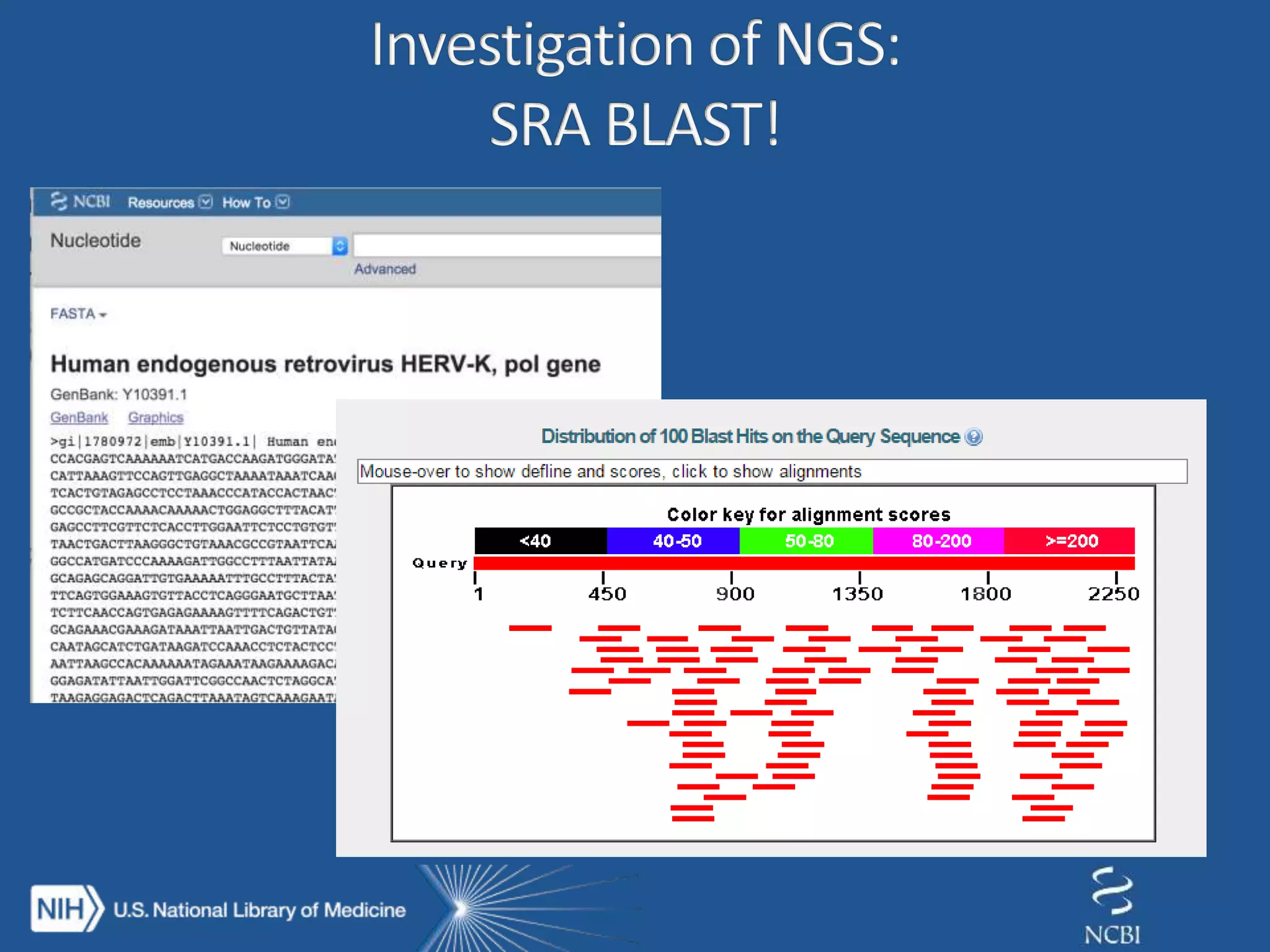
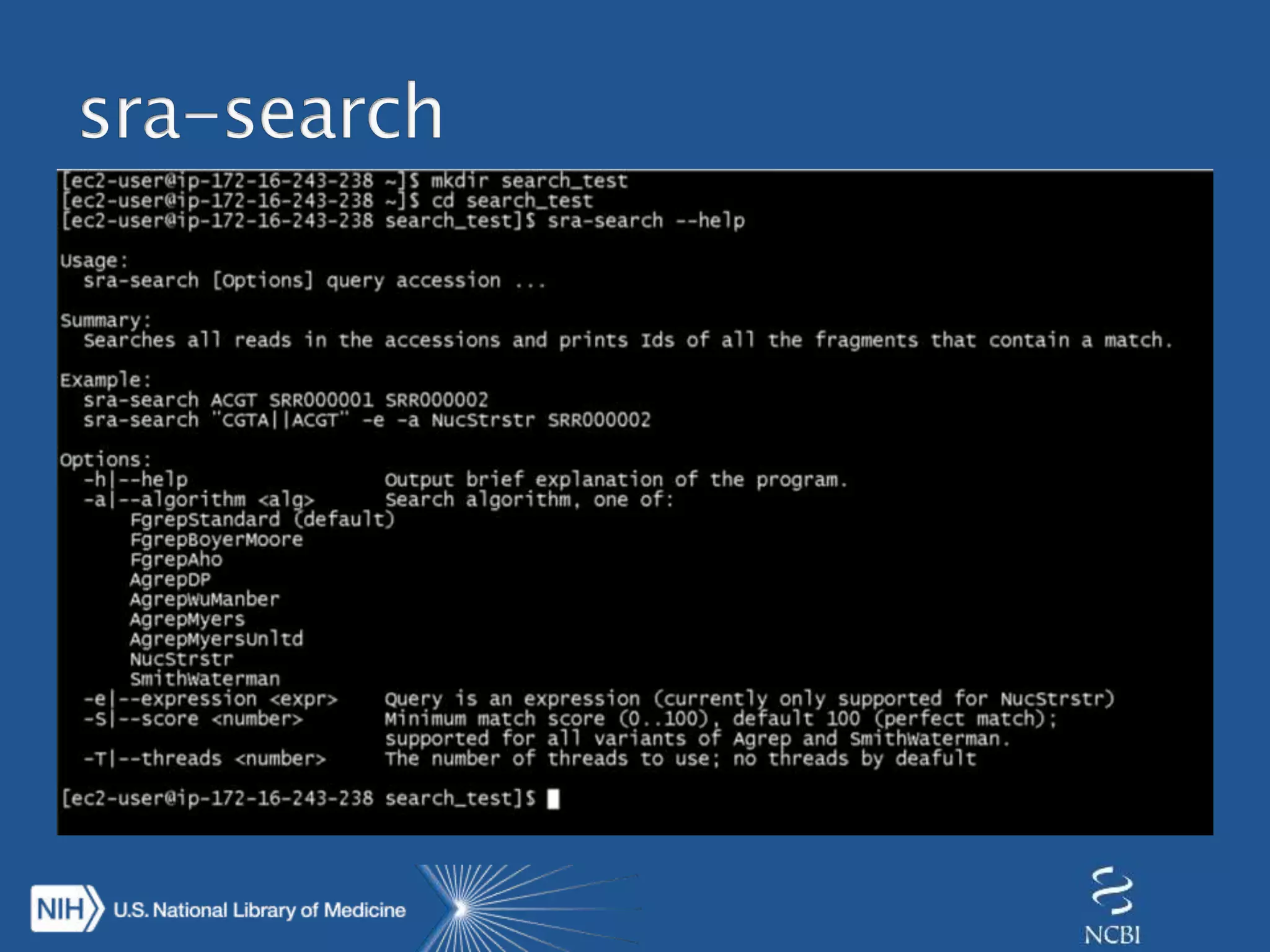
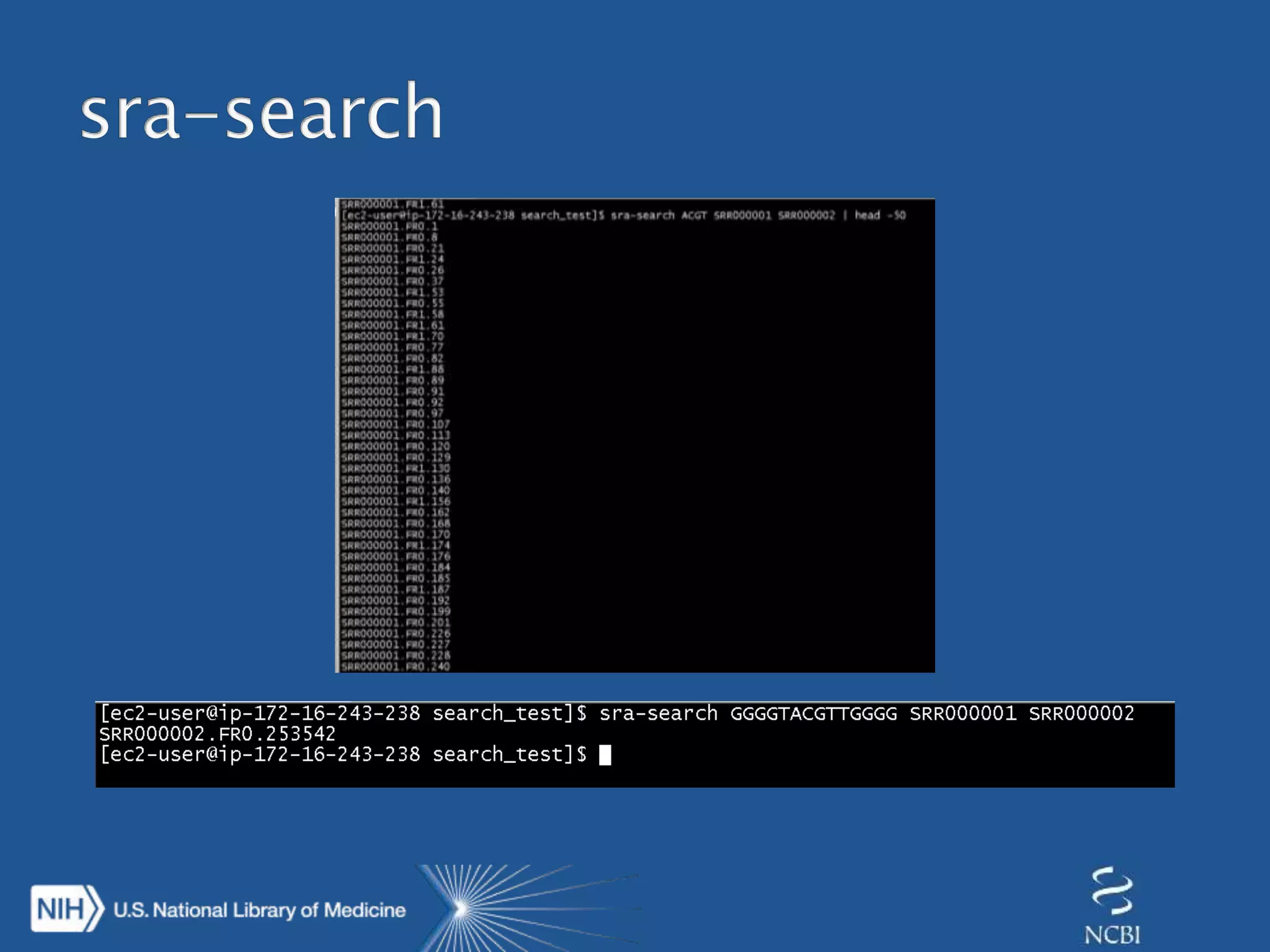
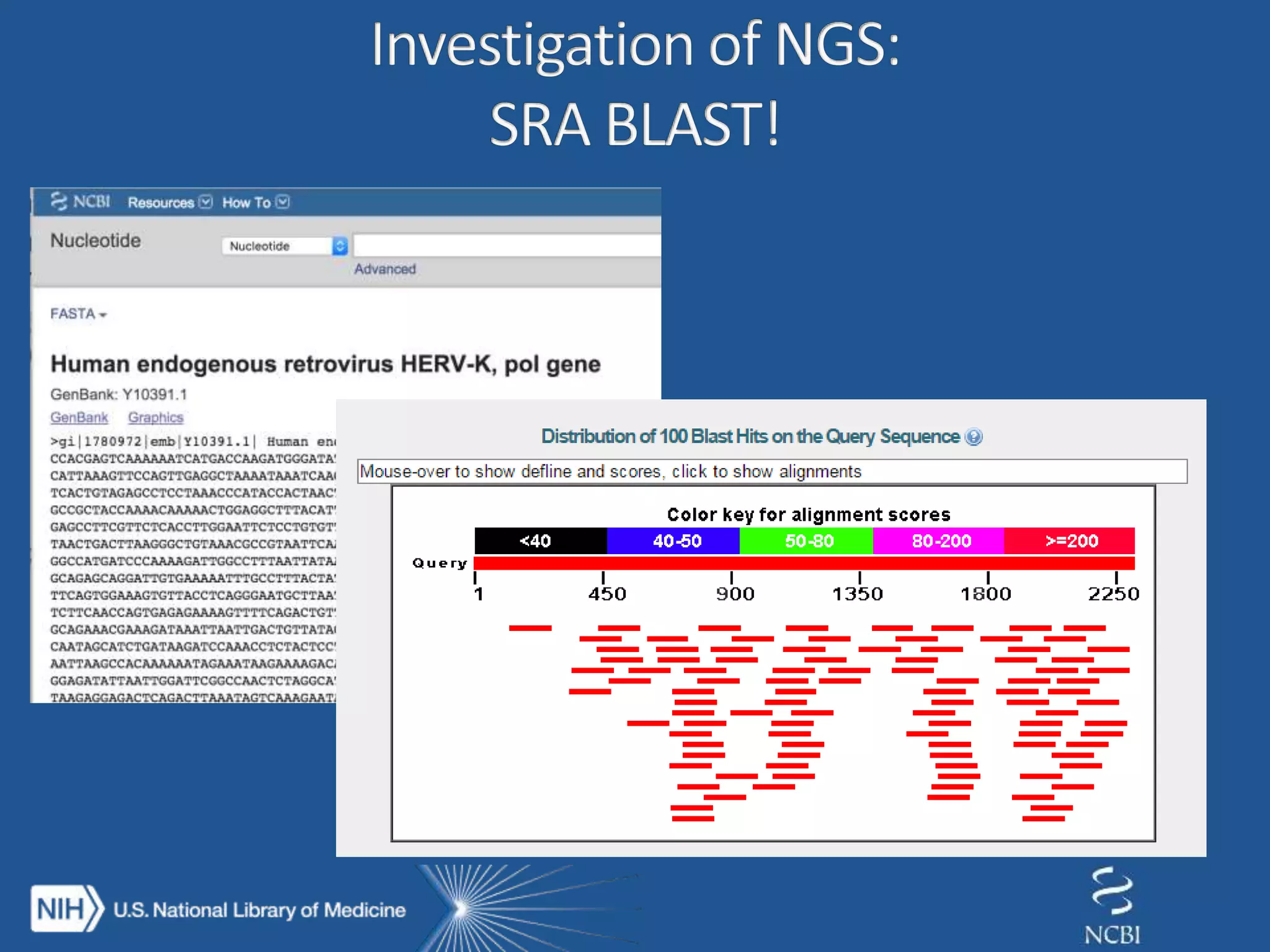
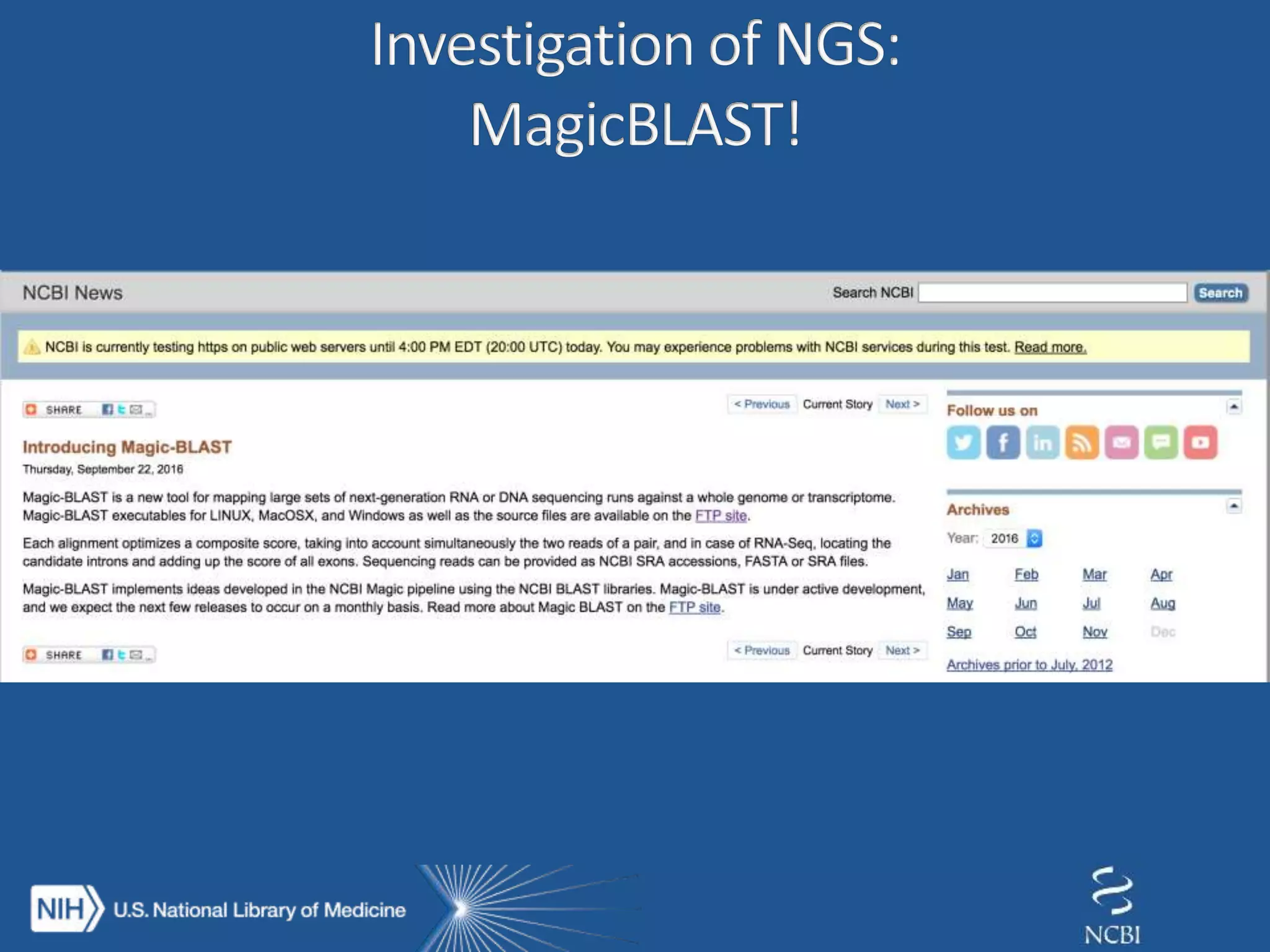
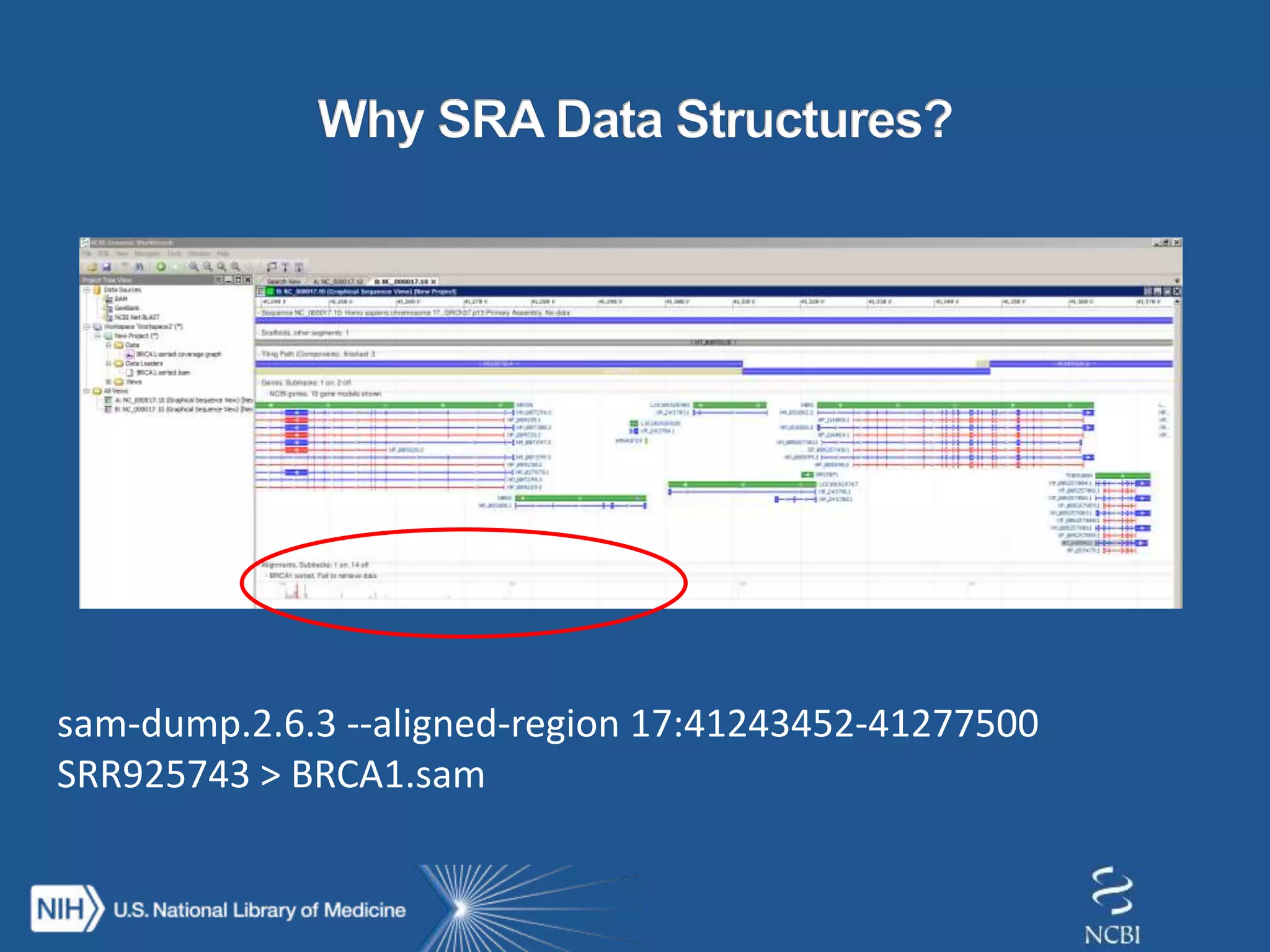
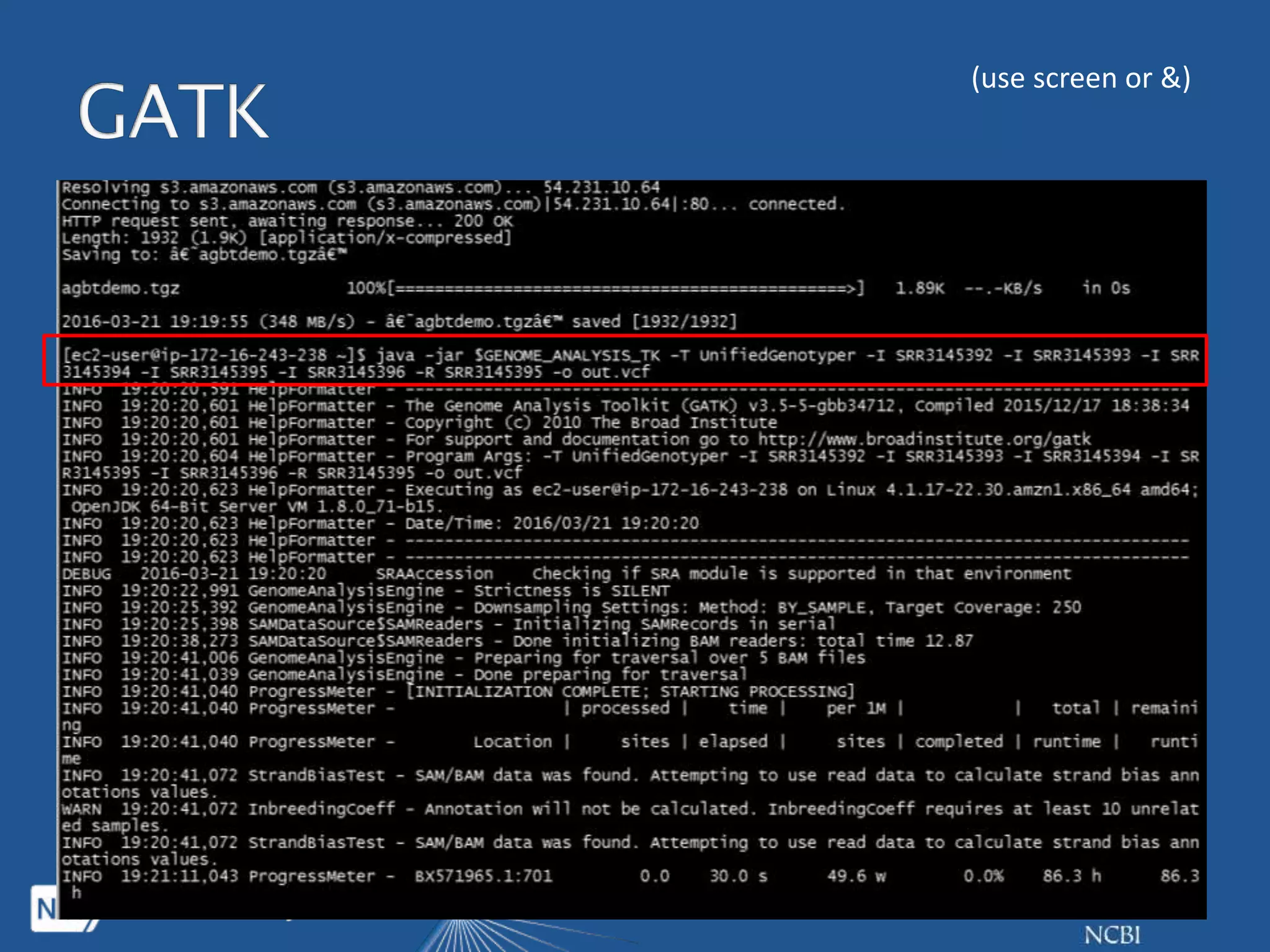
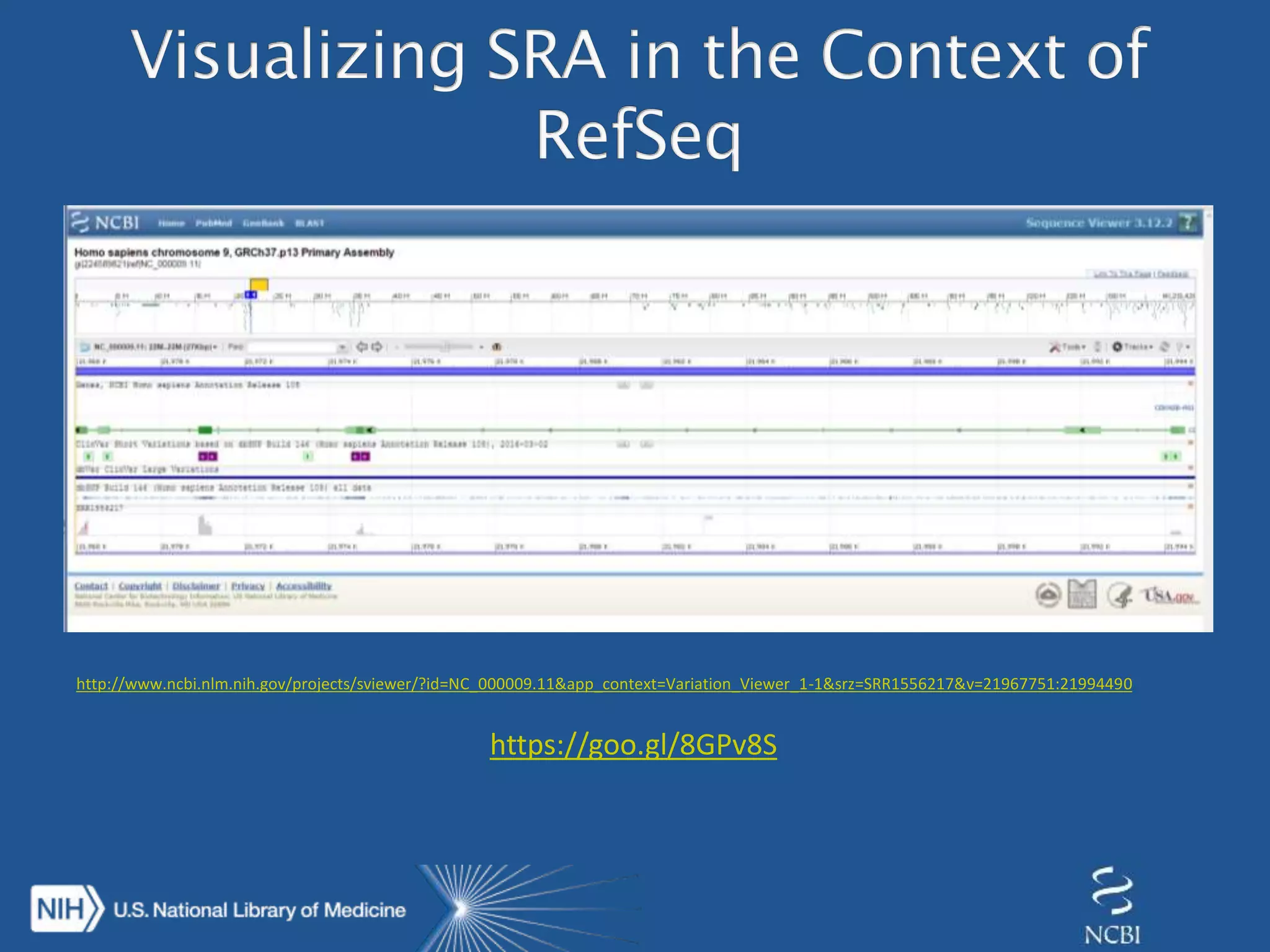
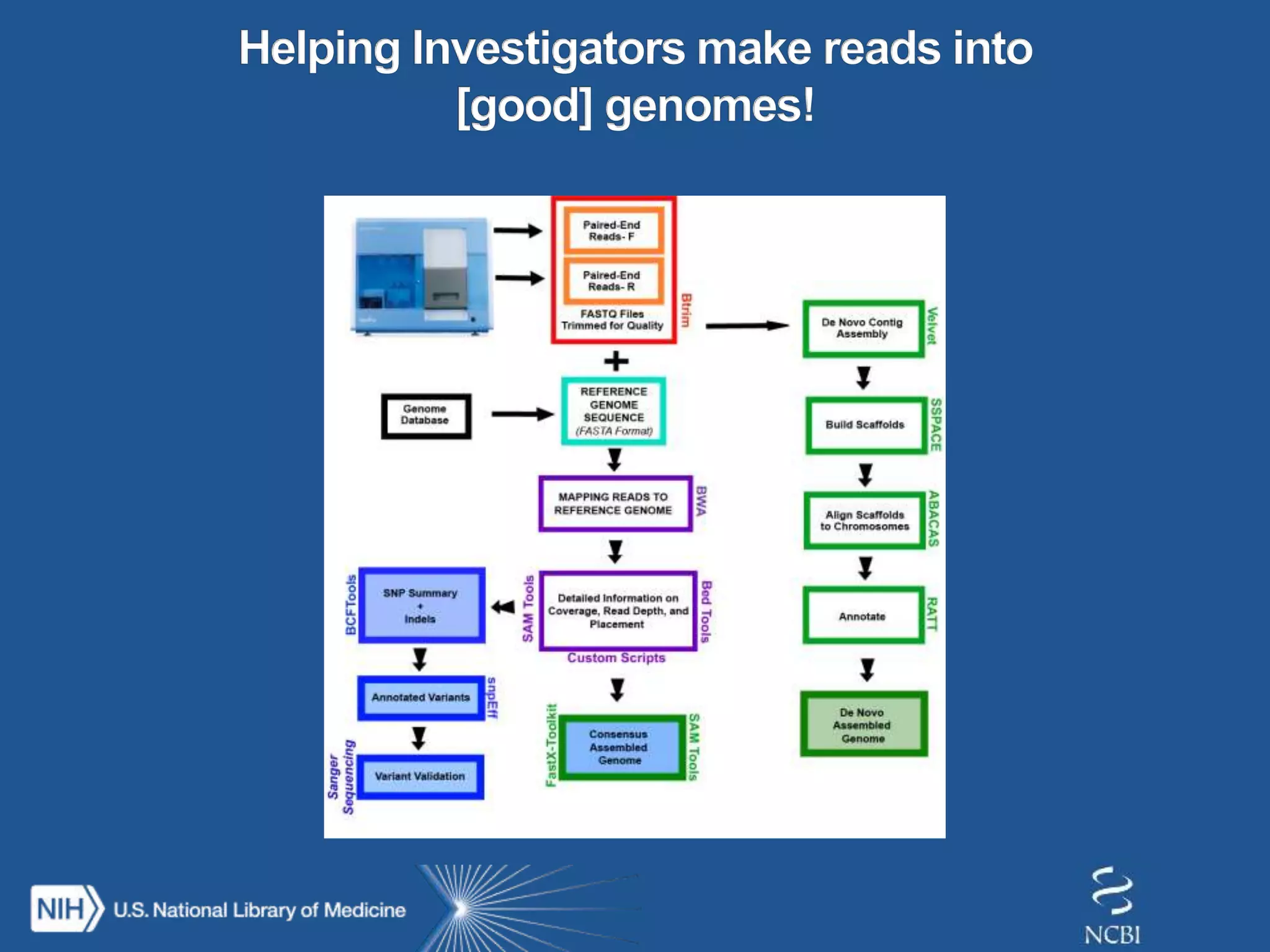
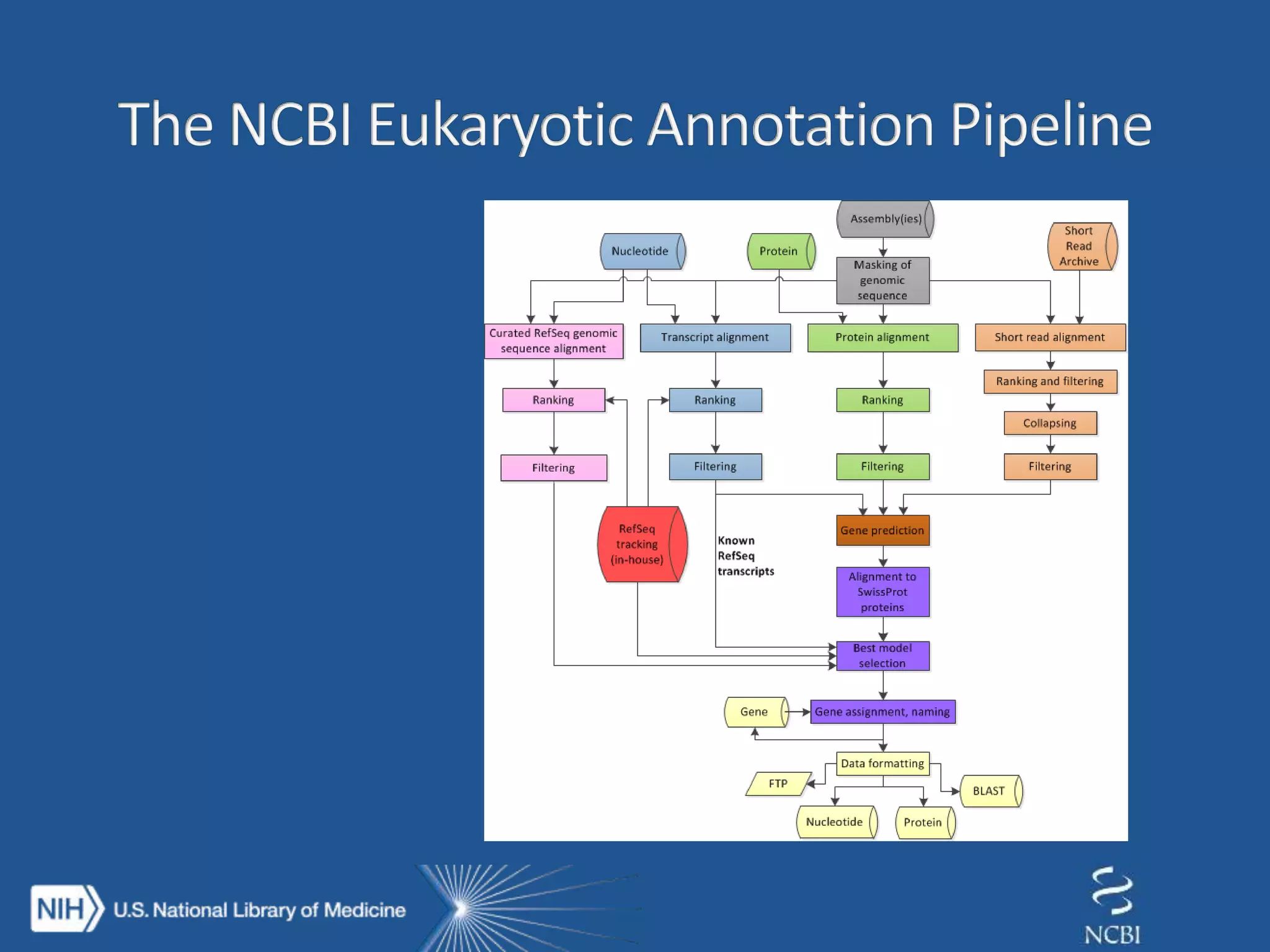
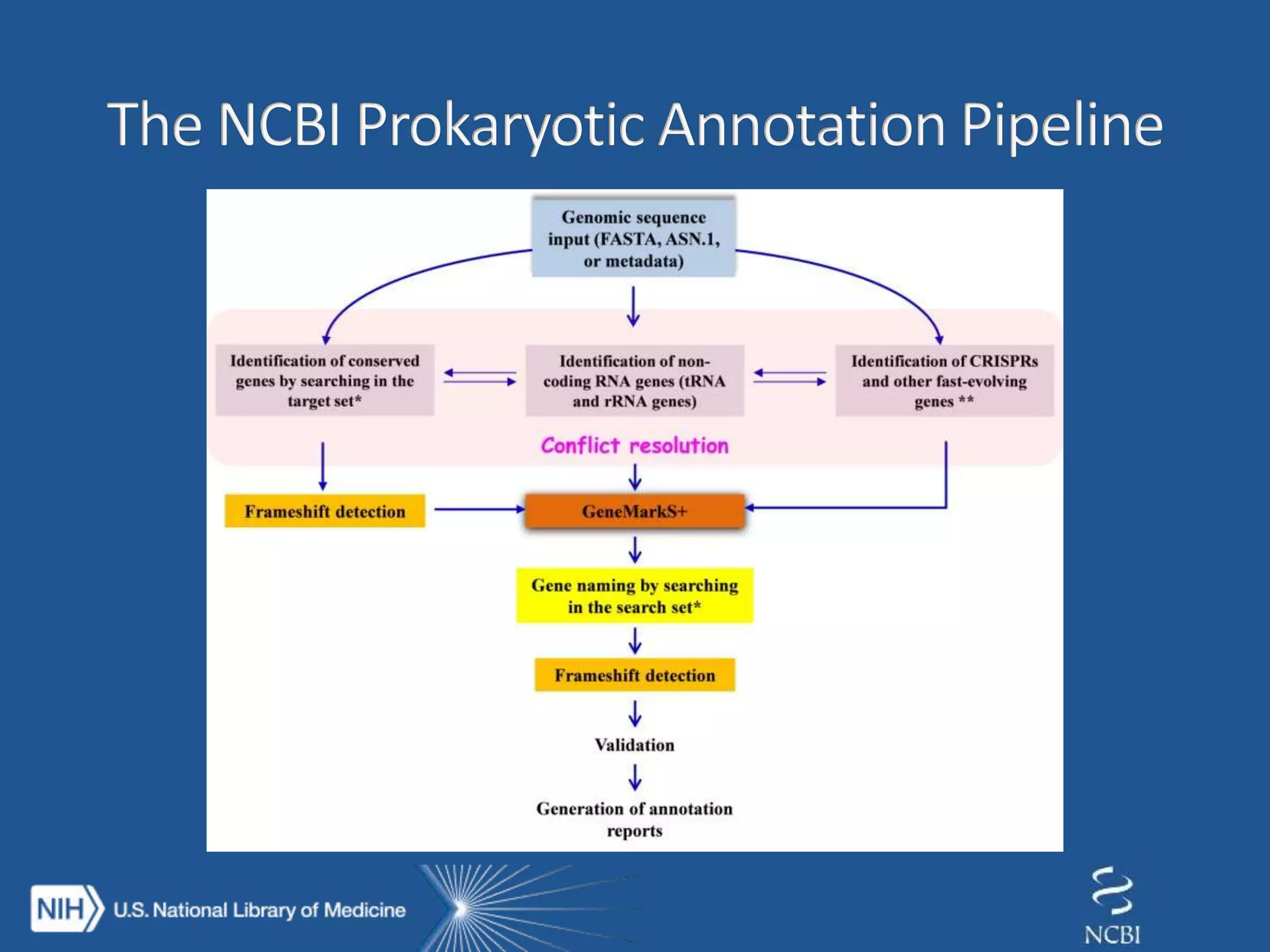
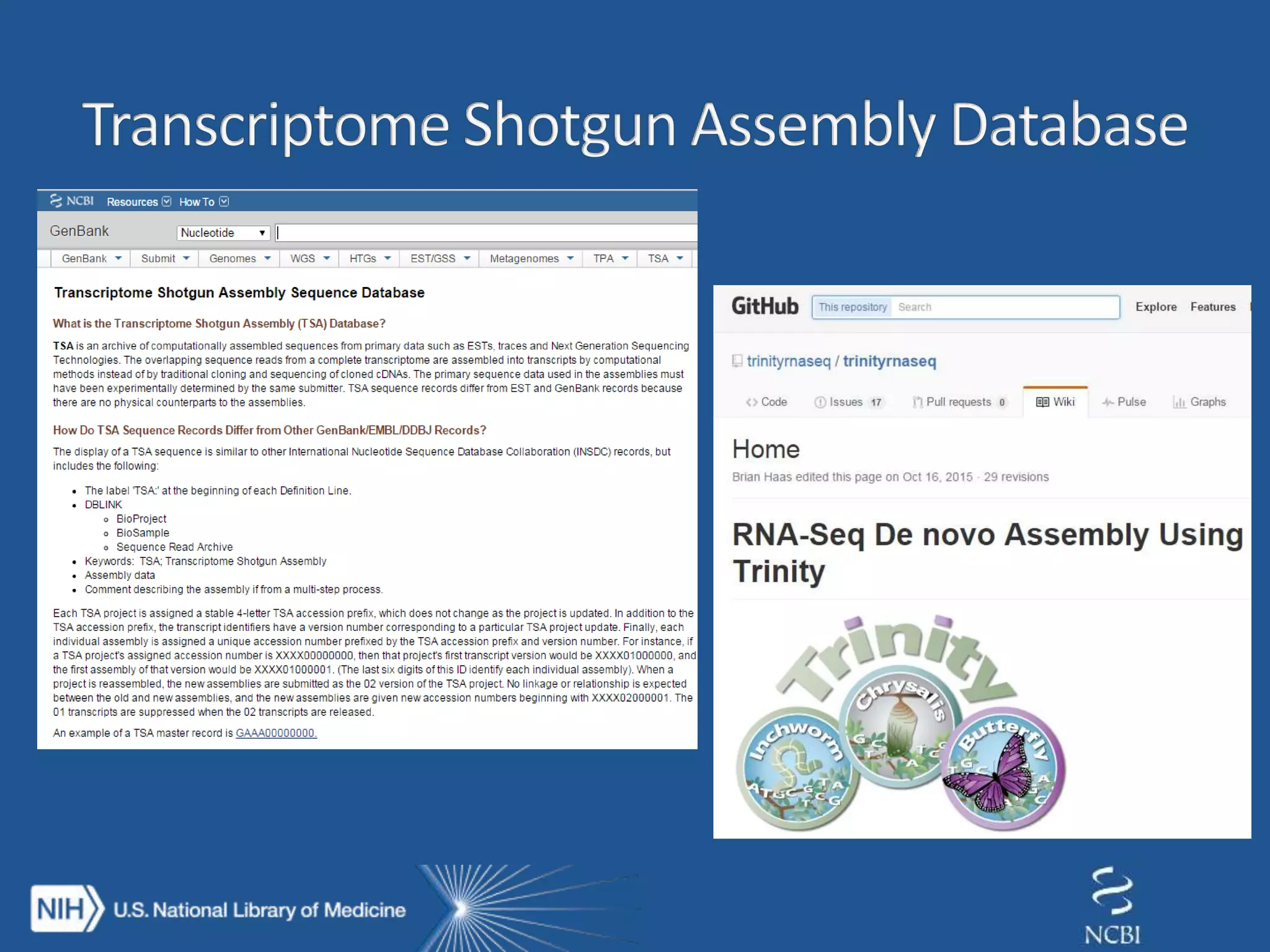
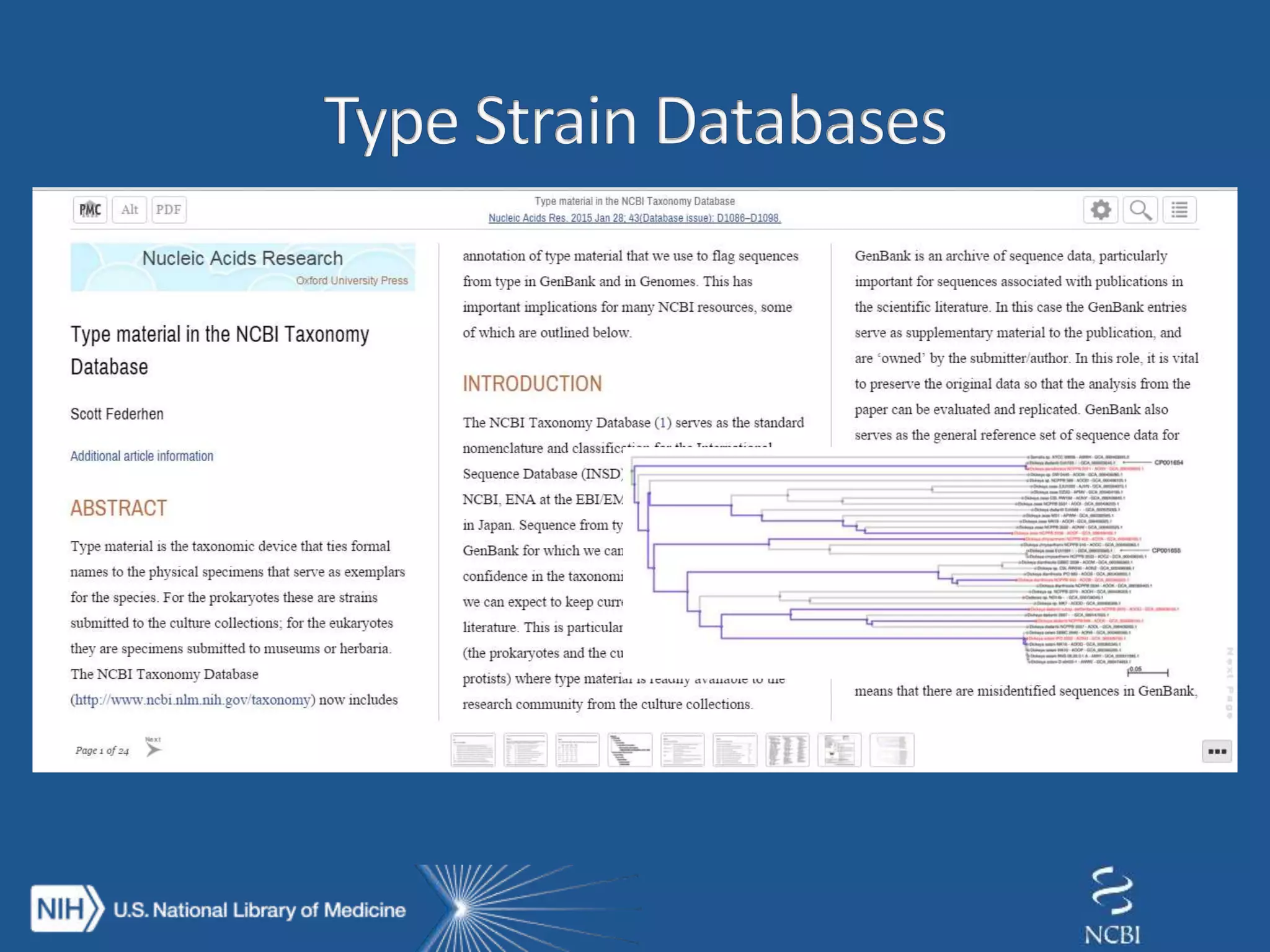
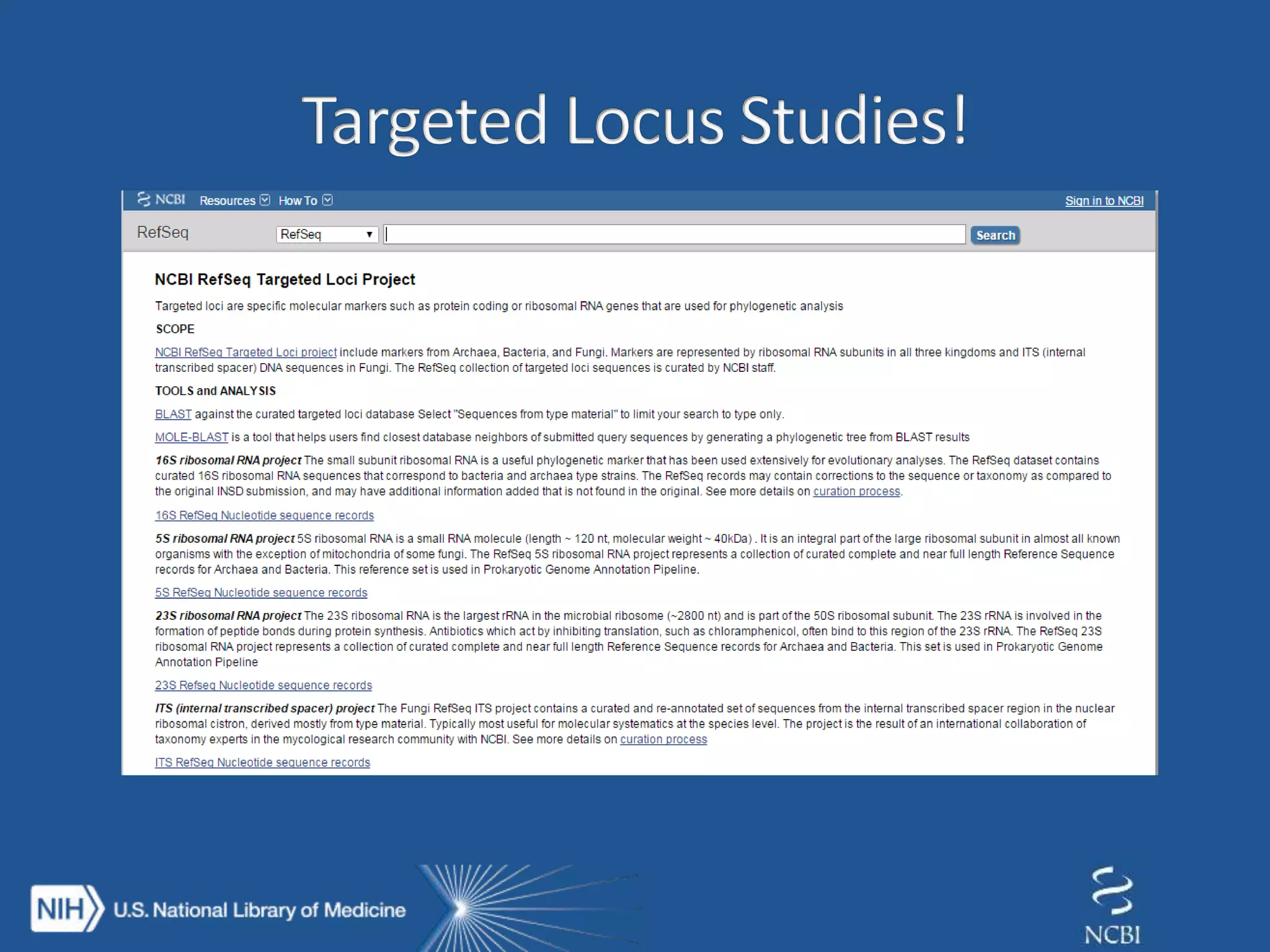
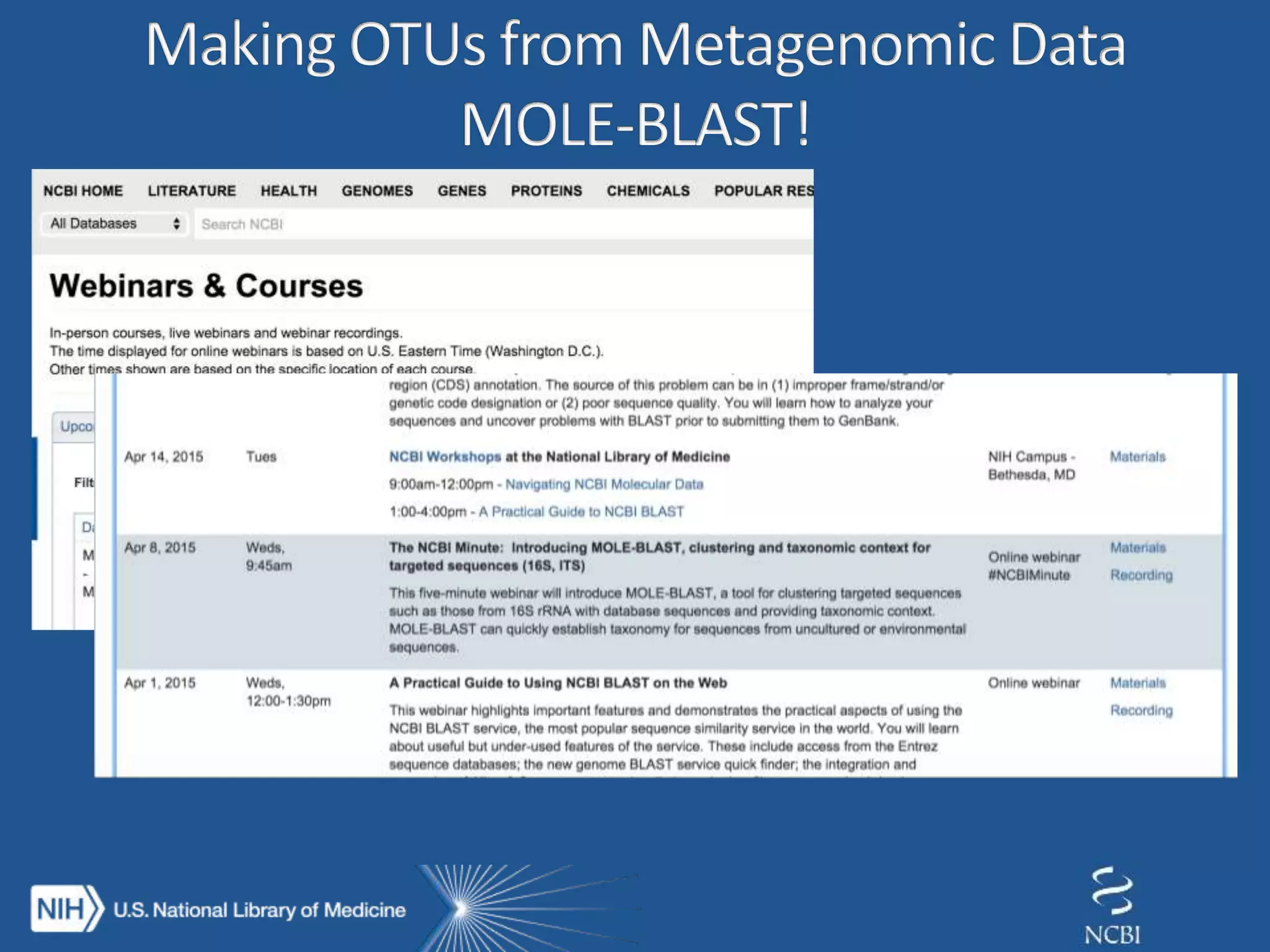
This document summarizes genomic variation and individual genome sequencing. It discusses the rising number of subjects with genome sequences available, from 14,201 in 2007 to over 1 million in 2015. It also provides information about genomic analysis tools from NCBI including the SAM format, genome browsers, E-Utilities, Entrez Direct, and upcoming APIs. Finally, it advertises upcoming hackathons in January and March for developing tools and apps to work with NCBI genomic data and APIs.





![62
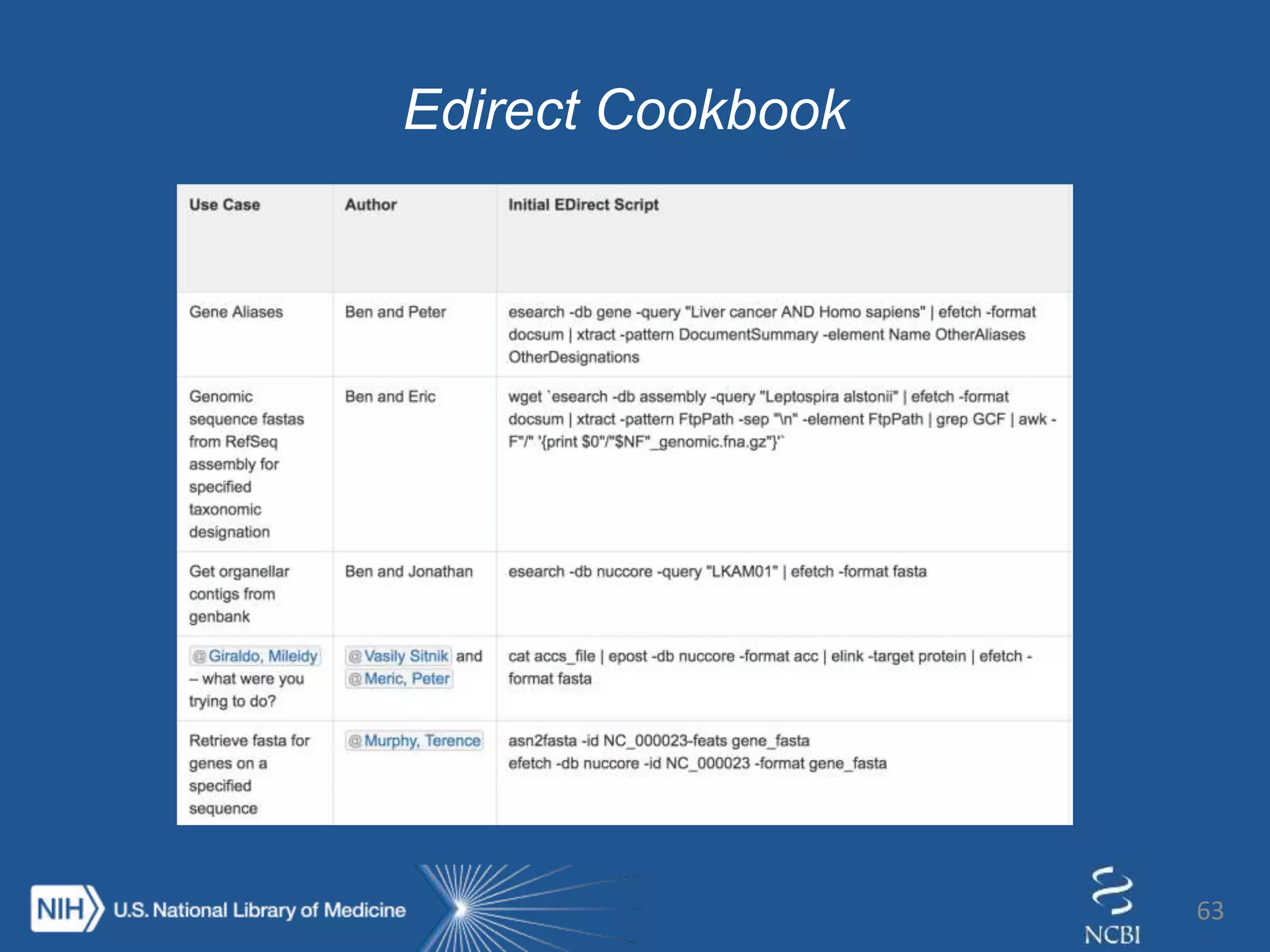
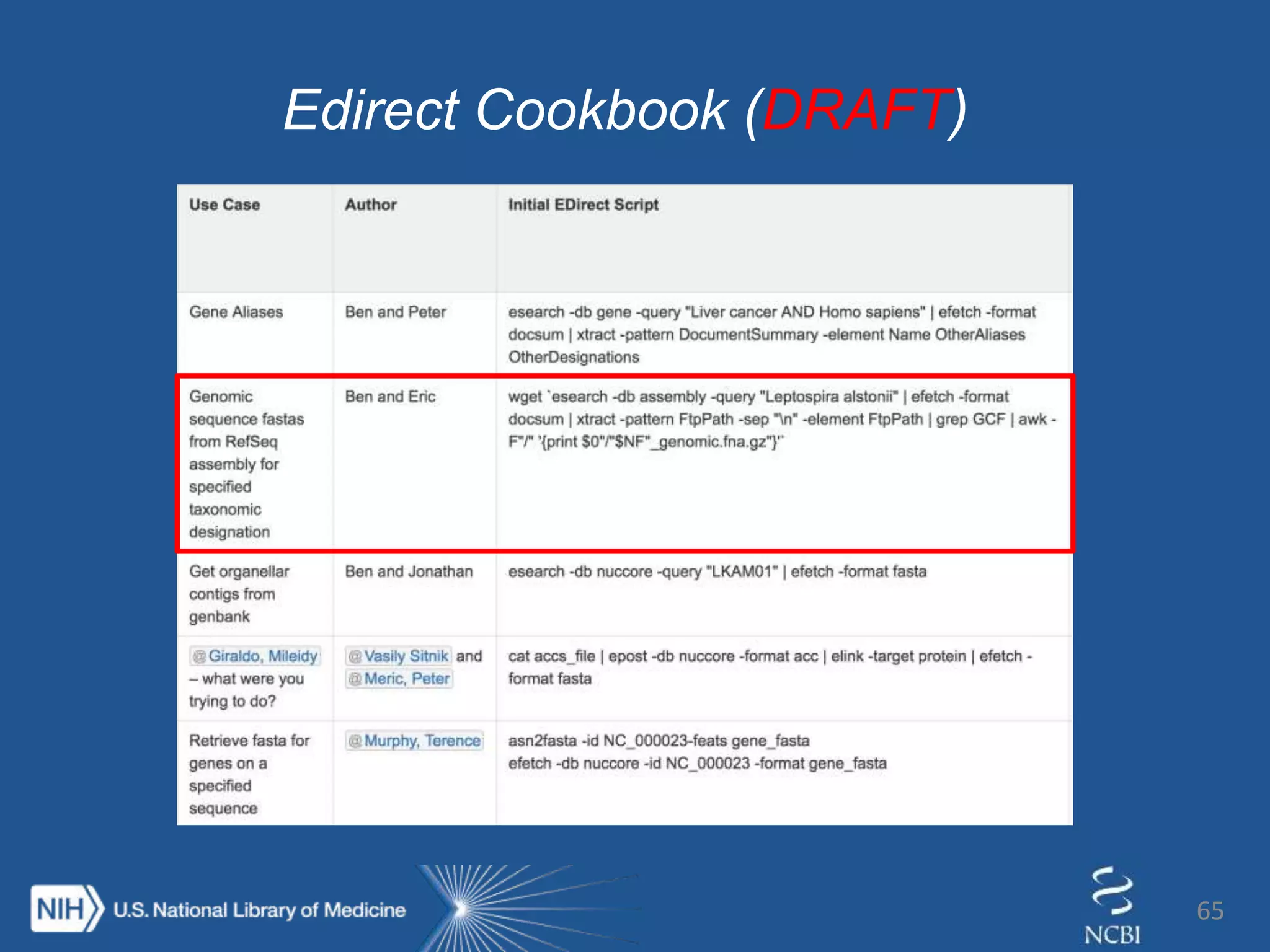
Introducing… Entrez Direct
The E-utilities on the UNIX
command line
esearch –db gene –query “foxp2[gene]
AND human[orgn]” |
elink –target protein –name
gene_protein_refseq |
efetch –format fasta
ftp.ncbi.nlm.nih.gov/entrez/entrezdirect/](https://image.slidesharecdn.com/umcpcstalk11316v1-161221161036/75/Umcp-cs-talk_11_3_16_v1-62-2048.jpg)