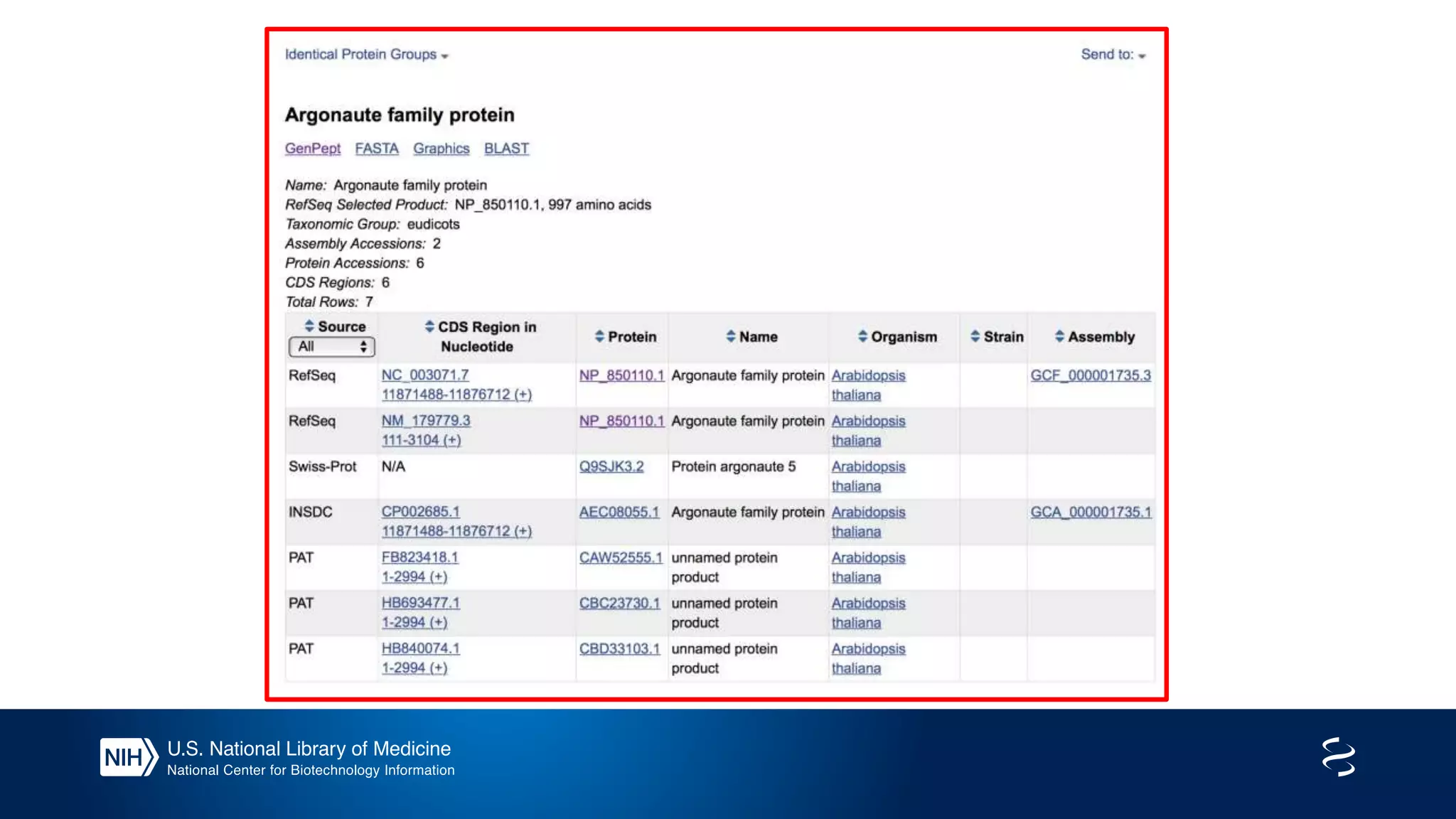
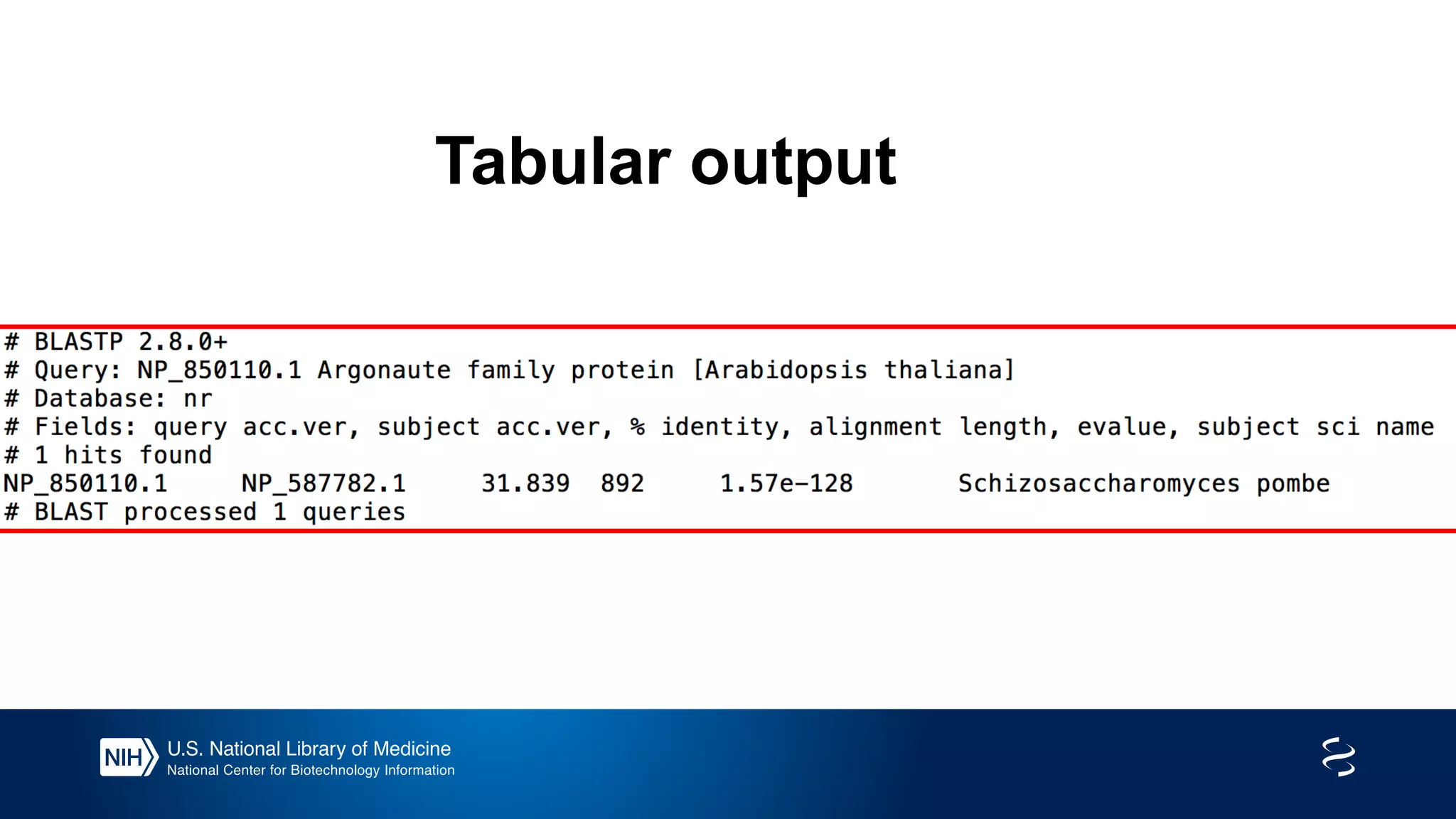
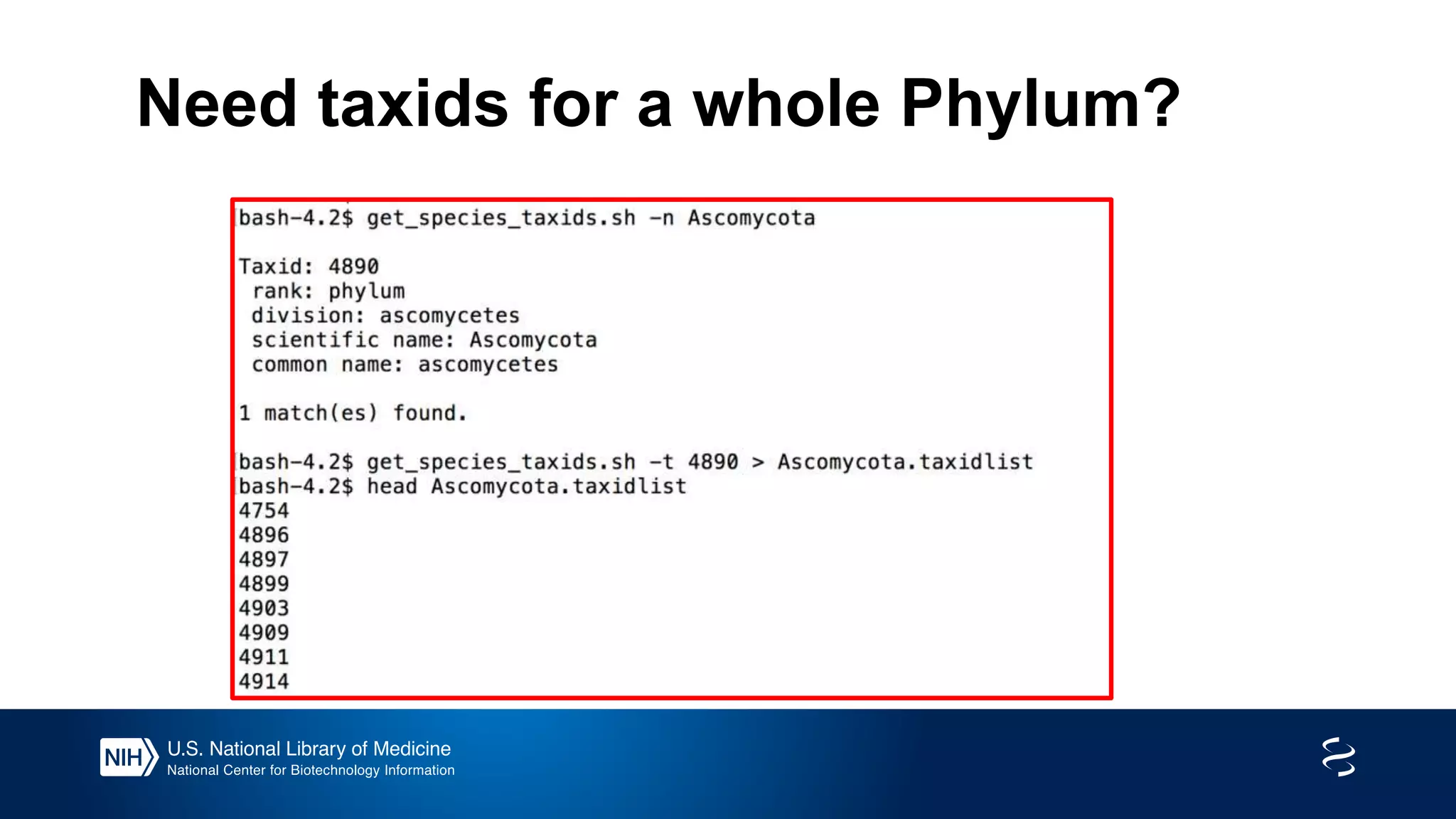
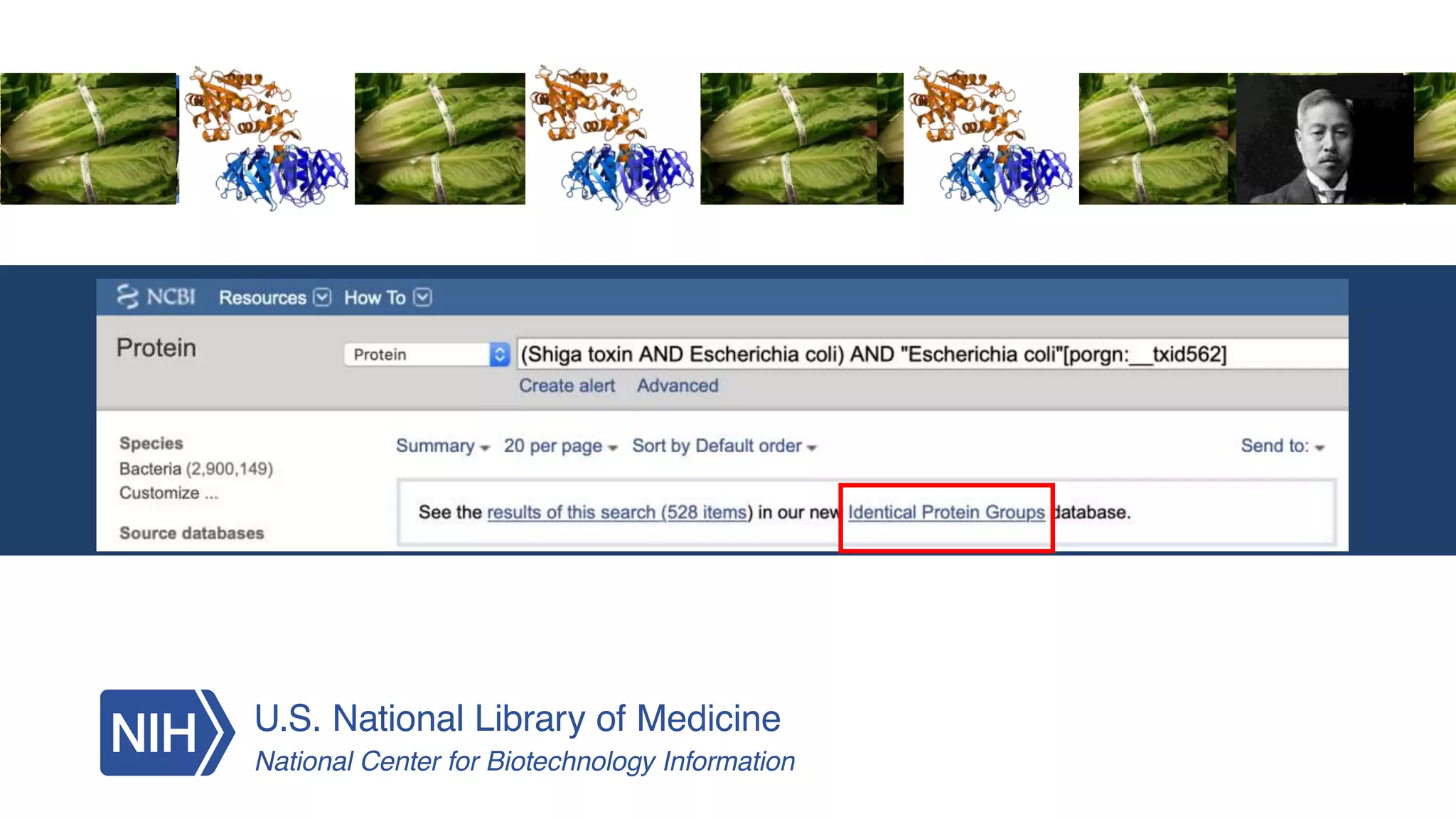
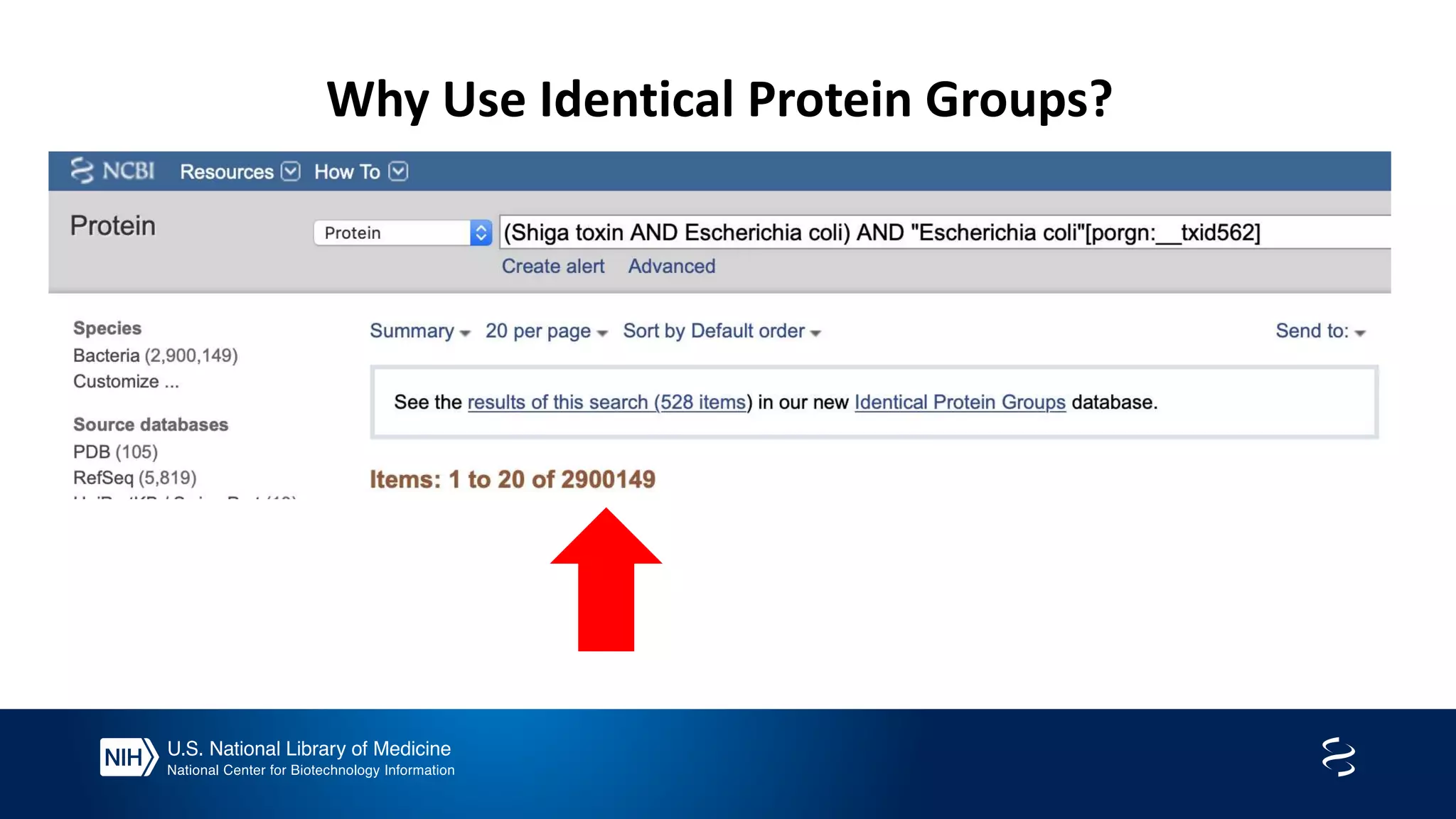
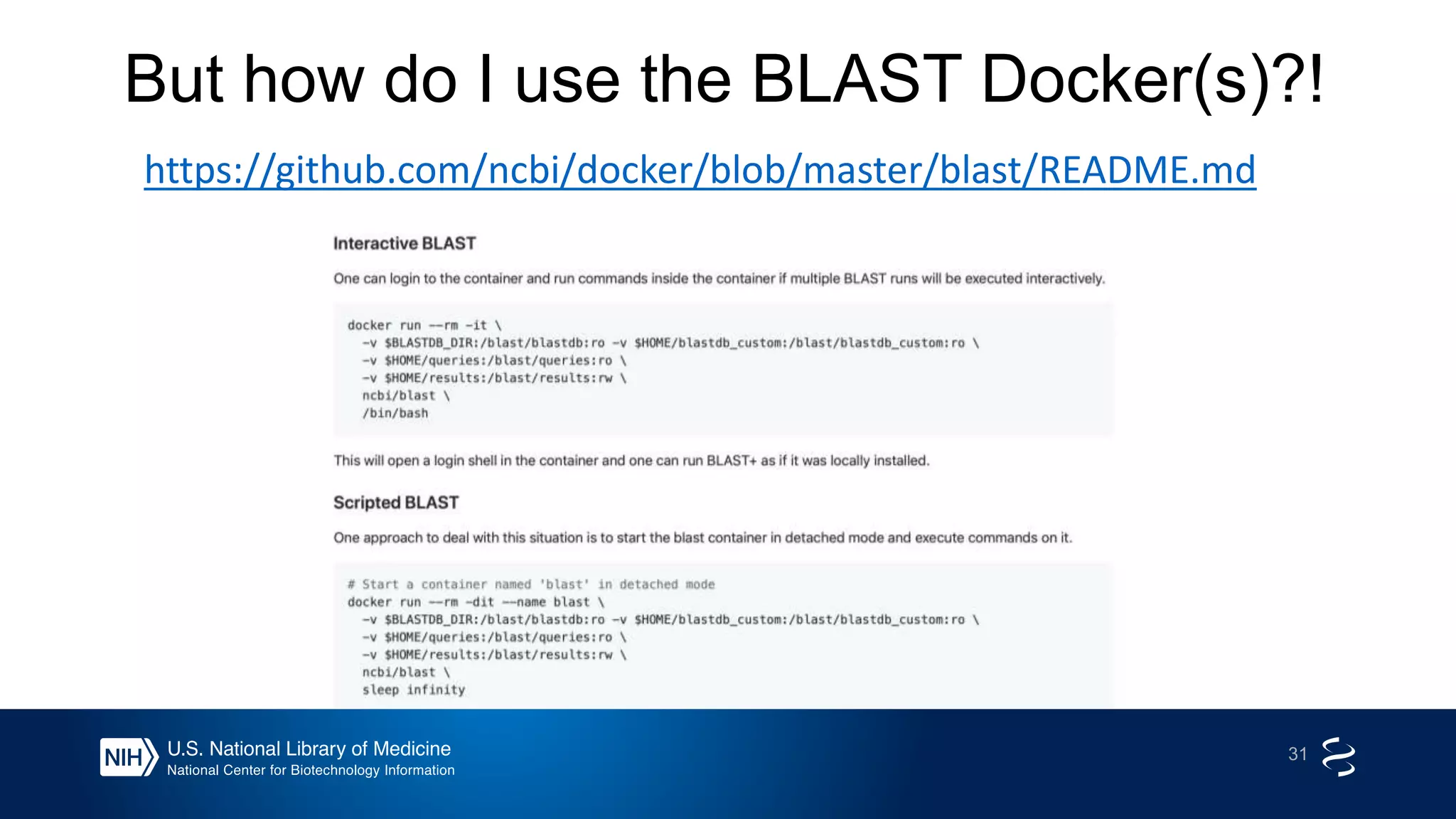
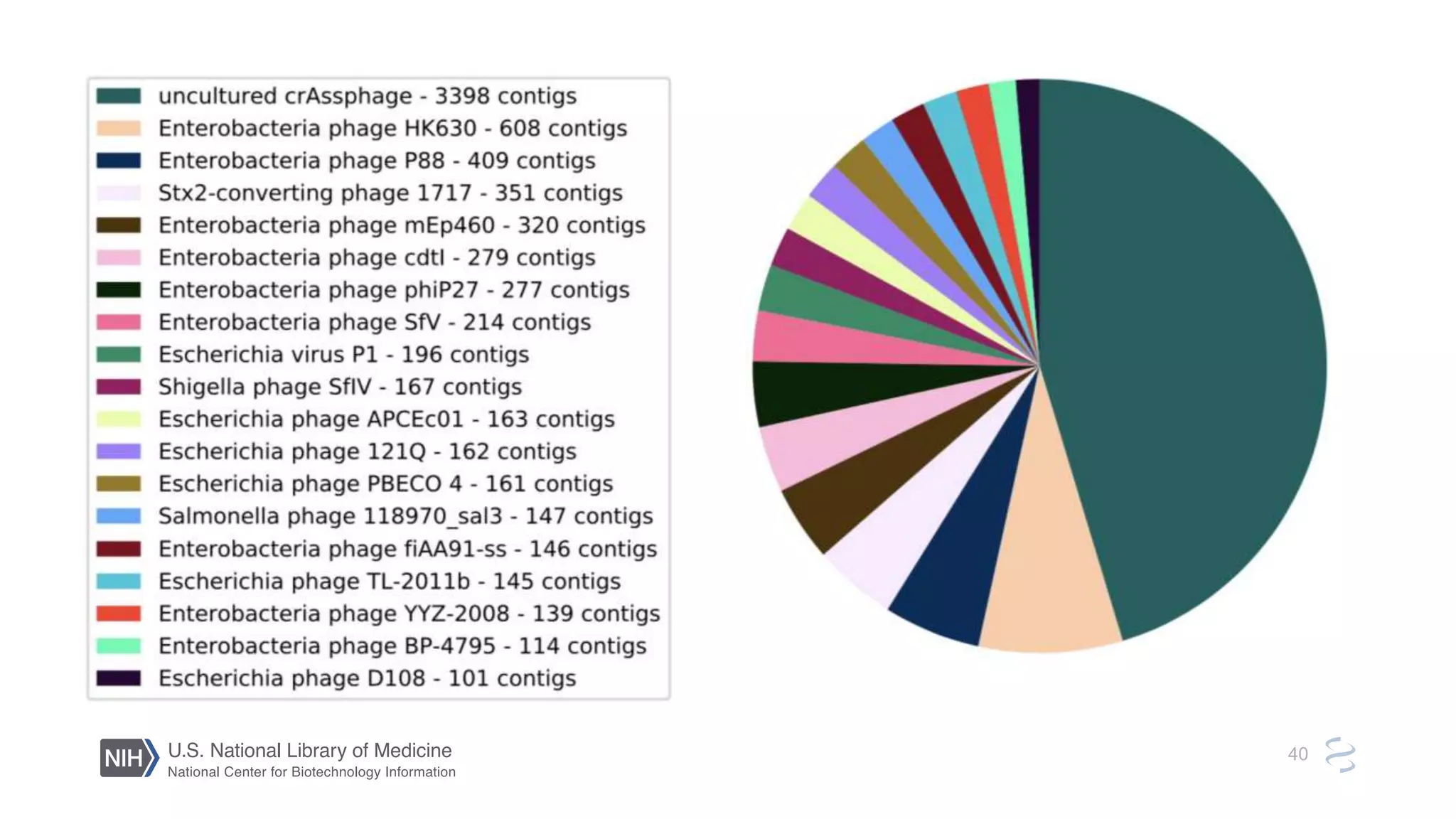
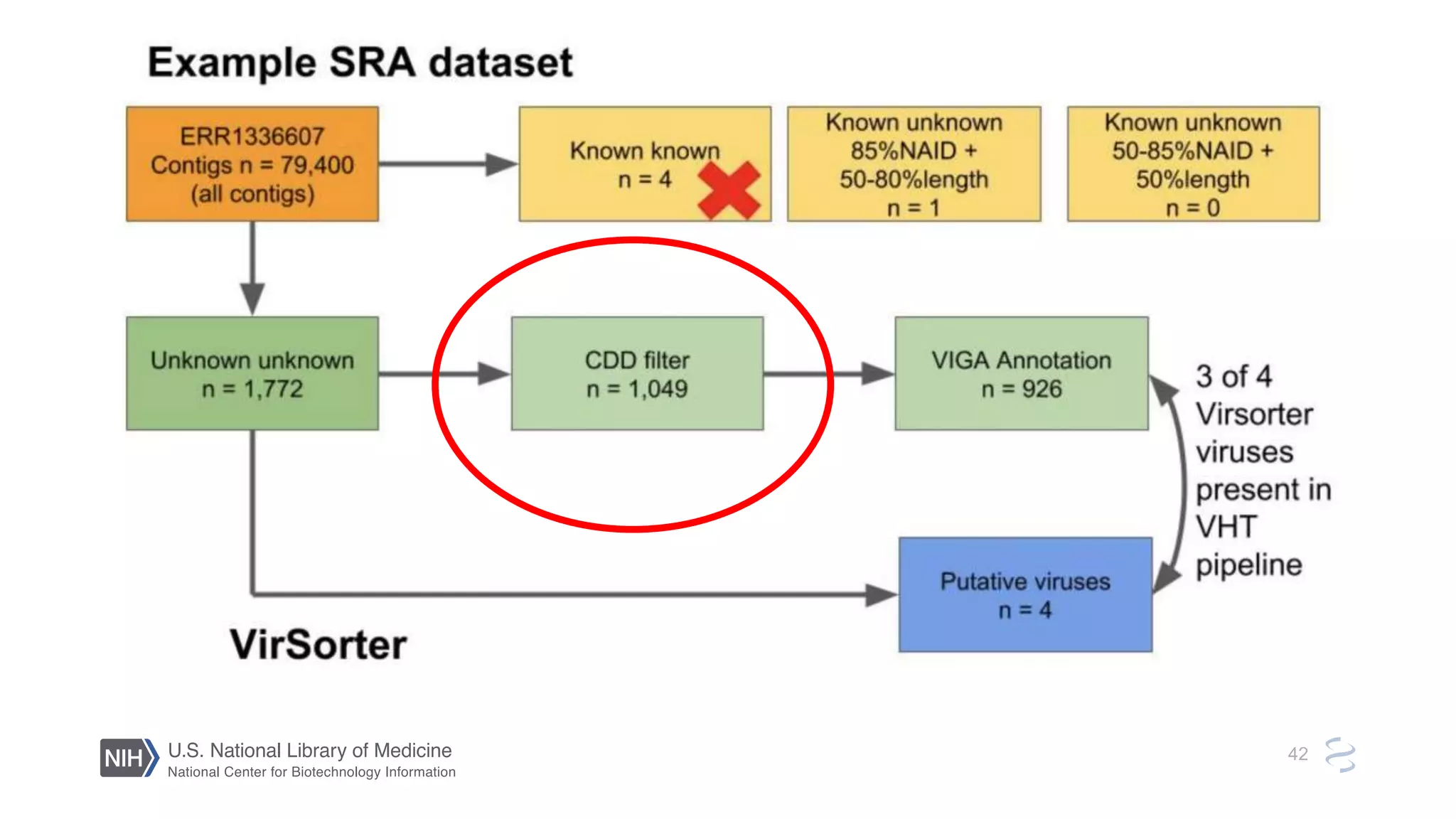
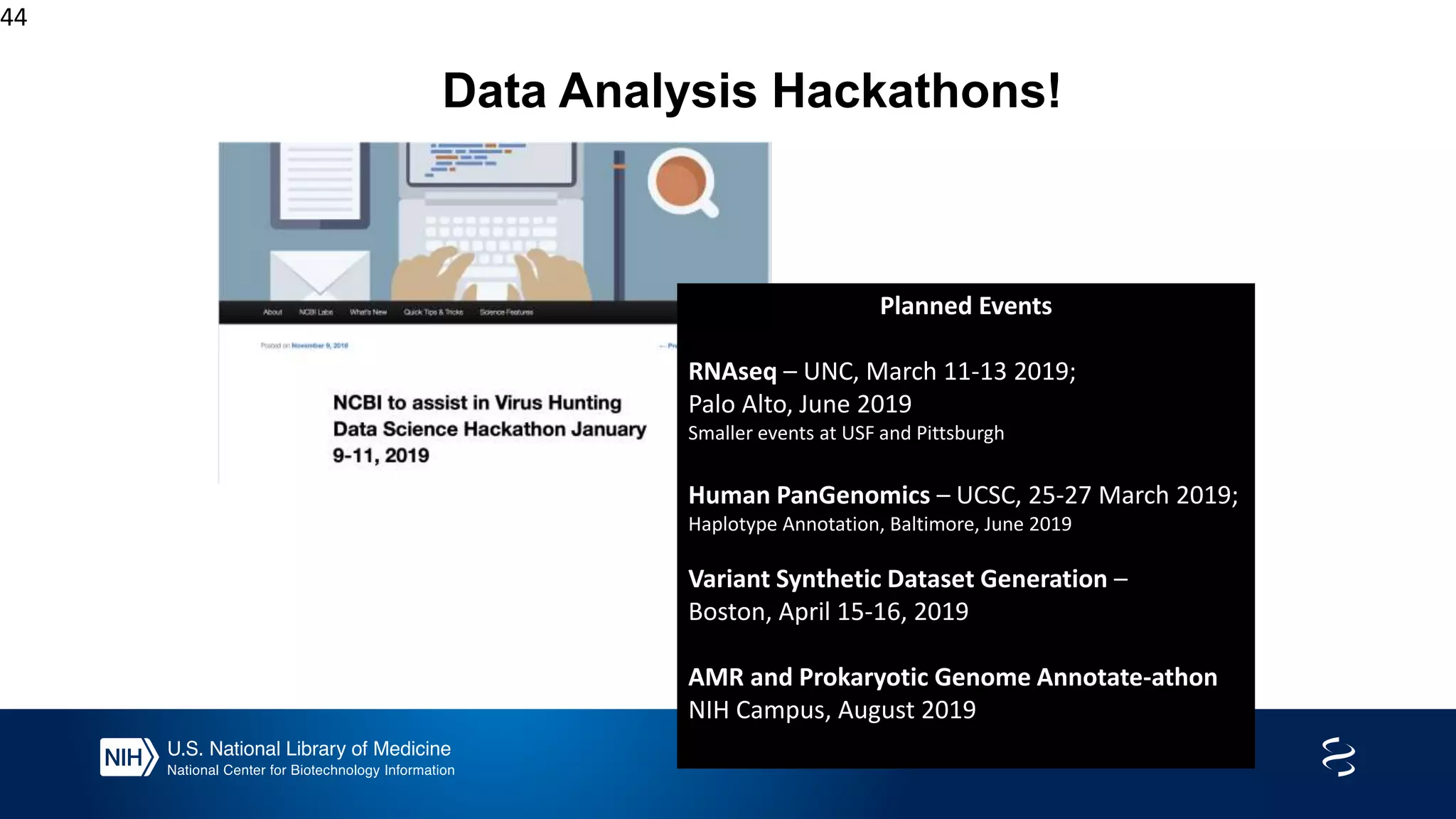
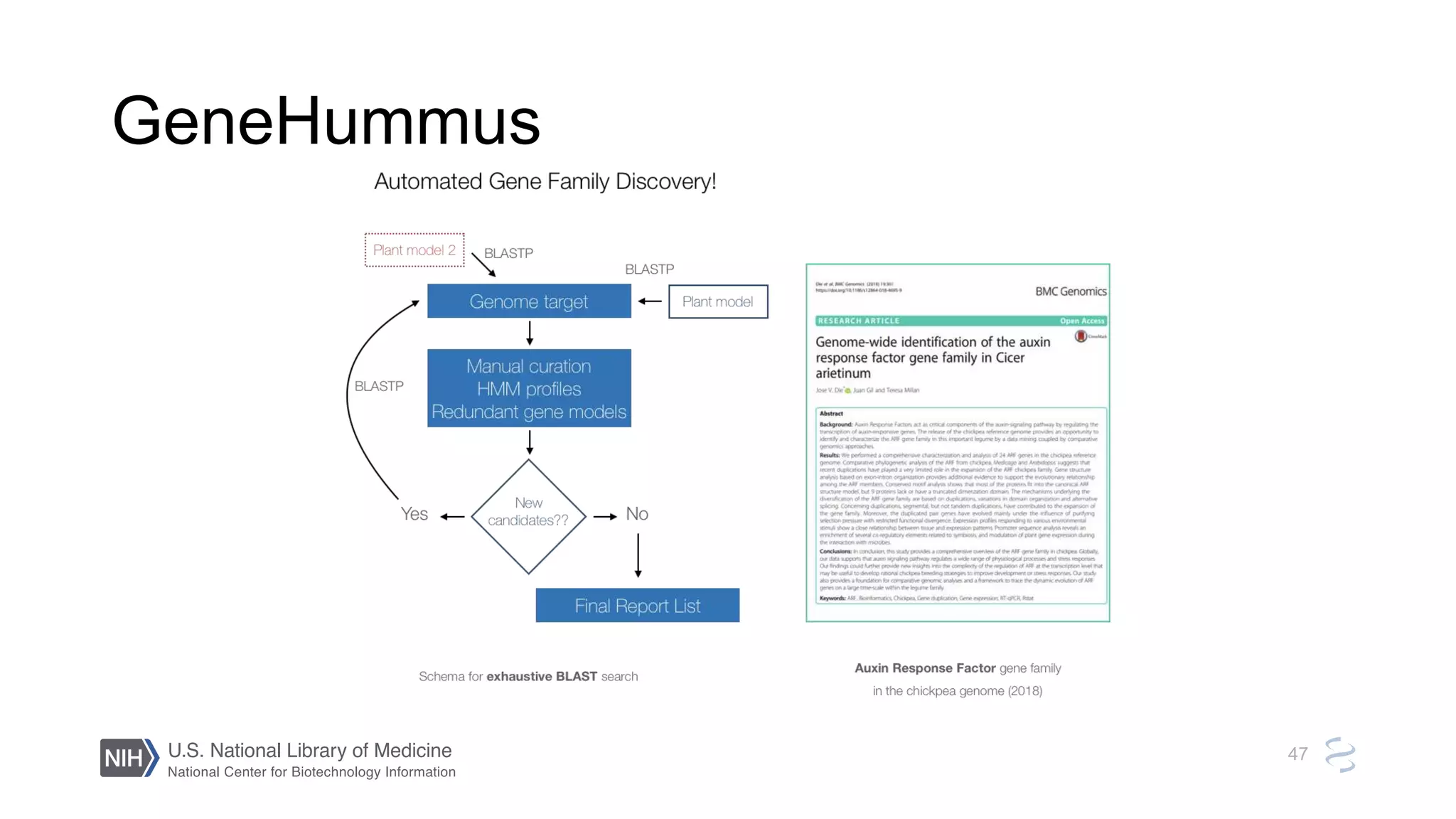
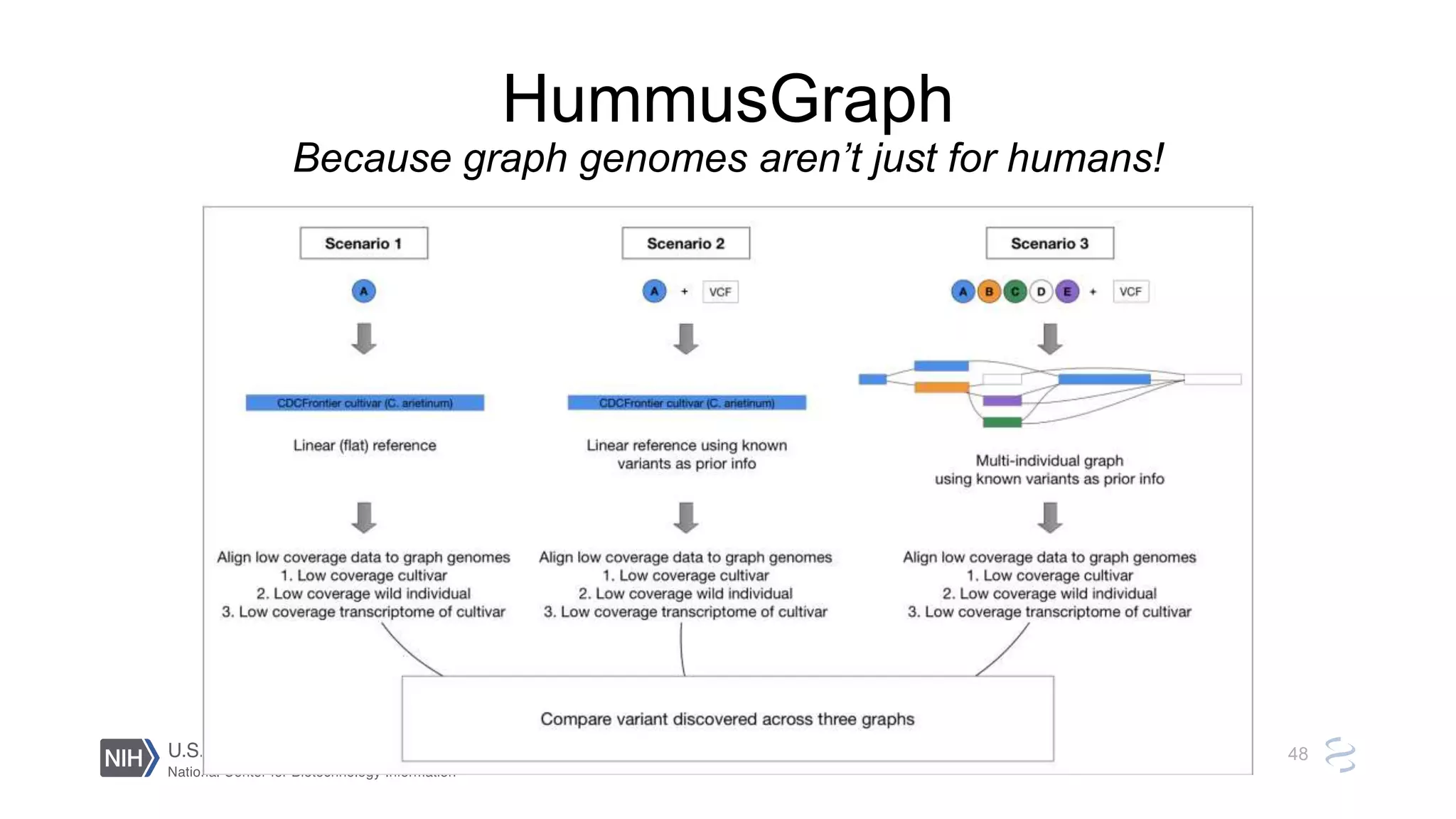
This document summarizes new features in NCBI BLAST that allow for more targeted searches based on taxonomy. It provides examples of using BLAST to search for Argonaute proteins in S. pombe, Ascomycota fungi, and excluding Viridiplantae. Execution times for each search are included. The document also demonstrates using BLAST docker images to search for Shiga toxin proteins in E. coli without needing the entire nr database locally. It concludes by advertising upcoming NCBI hackathons.