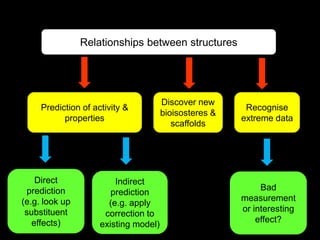
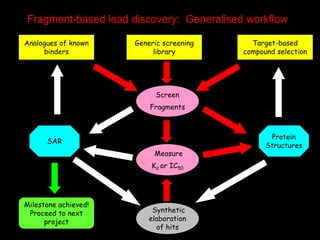
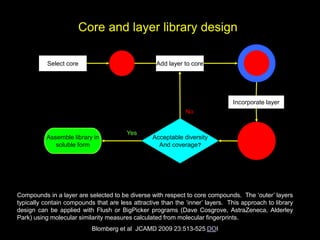
This document discusses molecular design for drug discovery. It outlines how molecular design can be used to control compound behavior through manipulation of molecular properties. Hypothesis-driven and prediction-driven approaches to molecular design are discussed. Relationship between structures, such as bioisosterism, are important for analyzing biological activity and physicochemical properties. Library design for screening is also covered, focusing on diversity, coverage, and neighborhood sampling of chemical space.




![TEP= [퐷푟푢푔푿,푡]푓푟푒푒 퐾푑
Target engagement potential (TEP)
A basis for pharmaceutical molecular design?
Design objectives
•Low Kdfor target(s)
•High (hopefully undetectable) Kdfor antitargets
•Ability to control[Drug(X,t)]free
Kenny, Leitão& MontanariJCAMD 2014 28:699-710 DOI](https://image.slidesharecdn.com/pwkuctoct2014-141024021445-conversion-gate02/85/UCT-Oct-2014-5-320.jpg)












![Leatherface molecule editor
From chain saw to Matched Molecular Pairs
c-[A;!R]
bnd 1 2
c-Br
cul 2
hyd 1 1
[nX2]1c([OH])cccc1
hyd 1 1
hyd 3 -1
bnd 2 3 2
Kenny & Sadowski Structure modification in chemical databases, Methods and Principles in Medicinal
Chemistry (Chemoinformatics in Drug Discovery 2005, 23, 271-285 DOI](https://image.slidesharecdn.com/pwkuctoct2014-141024021445-conversion-gate02/85/UCT-Oct-2014-19-320.jpg)