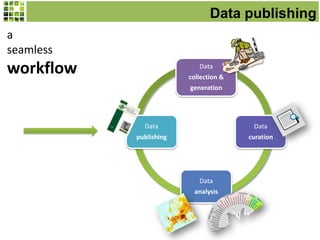
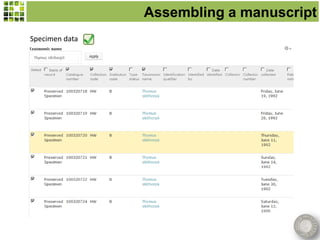
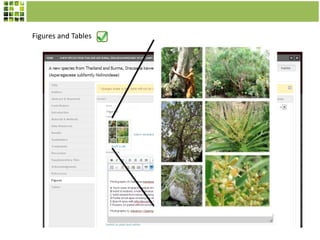
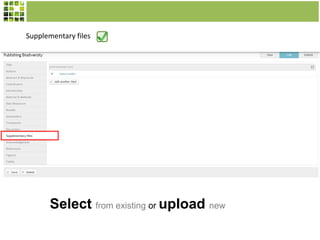
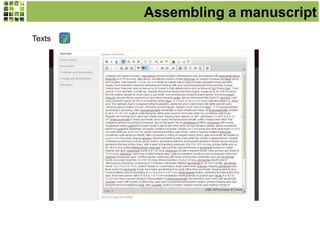
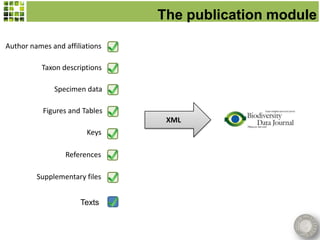
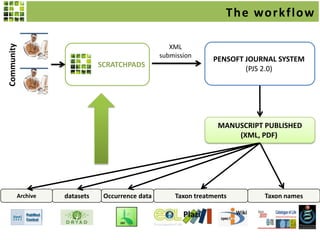
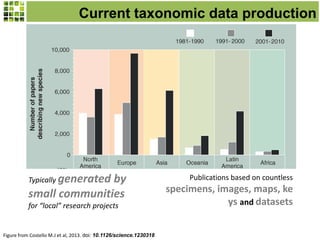
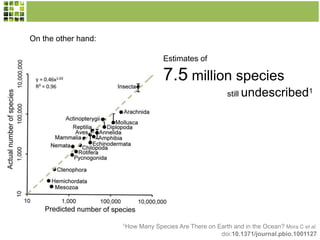
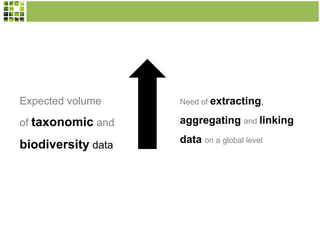
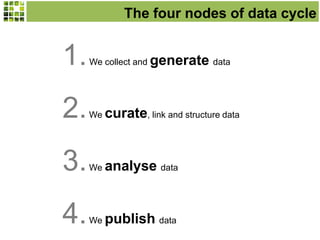
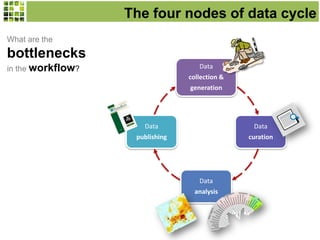
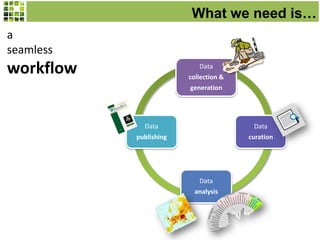
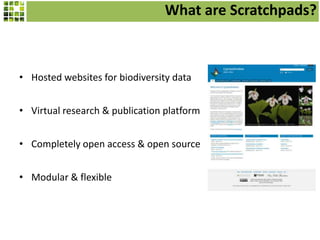
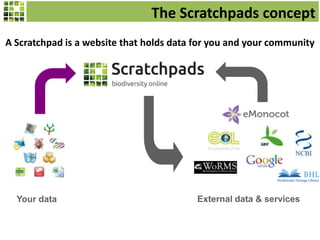
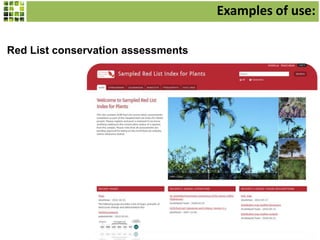
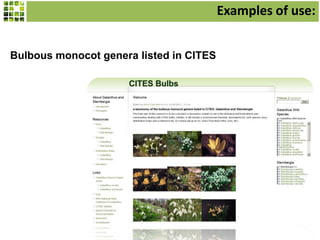
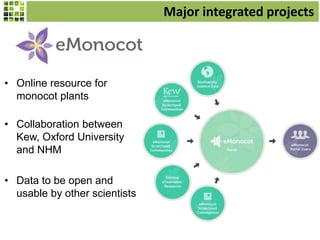
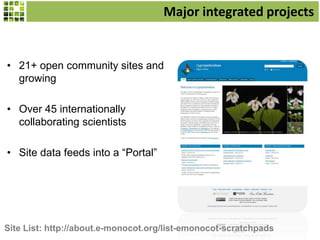
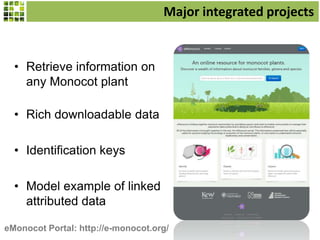
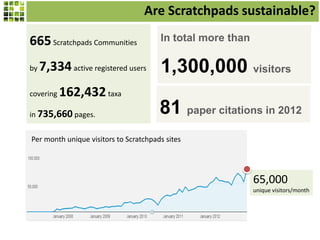
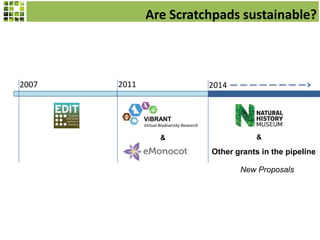
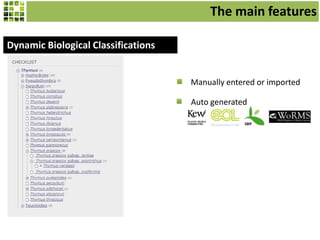
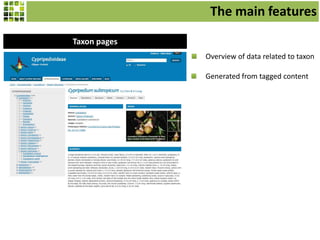
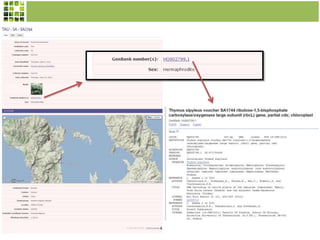
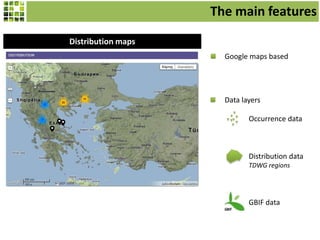
This document discusses Scratchpads, which are hosted websites that facilitate the digital curation and publication of taxonomic and biodiversity data. Scratchpads allow researchers to enter, link, and publish their data in a seamless, open access workflow. They support digital classifications, specimen records, bibliographies, distribution maps, and analytical tools. Major projects like e-Monocot use Scratchpads to aggregate data from over 45 researchers. Scratchpads currently involve over 7,000 users curating over 162,000 taxa. They integrate with services like GBIF, IUCN, and Plazi and allow publication in the open access Biodiversity Data Journal.












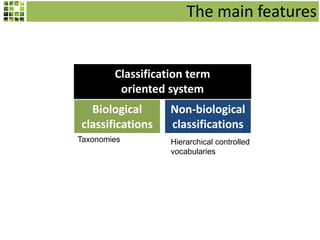


![The main features
Character matrices – Key construction
Quantitative or qualitative characters
Auto generation of keys
Taxon based matrices
[Specimens based character matrices]](https://image.slidesharecdn.com/scratchpad-2014-introduction-140224111002-phpapp01/85/Scratchpad-2014-introduction-37-320.jpg)