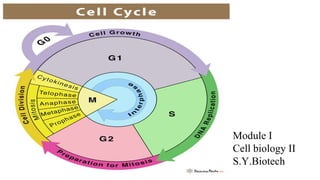
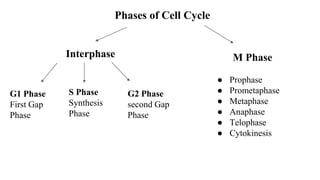
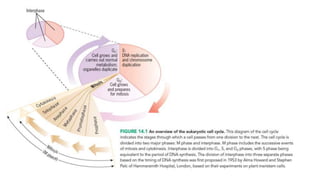
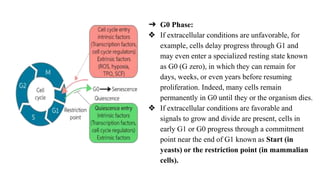
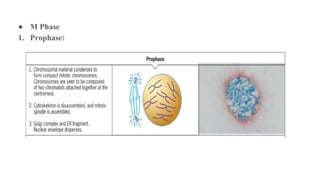
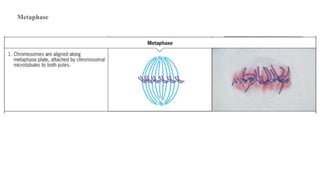
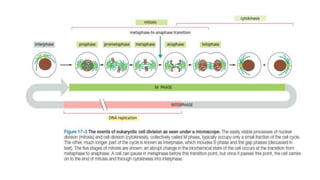
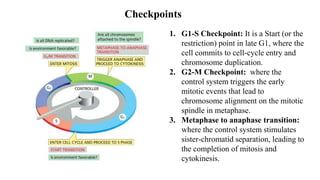
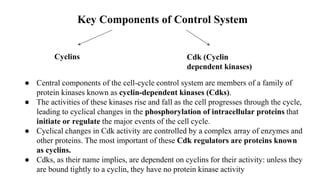
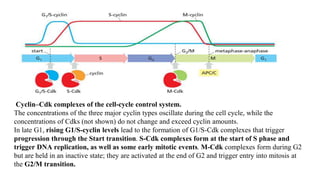
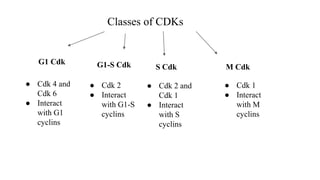
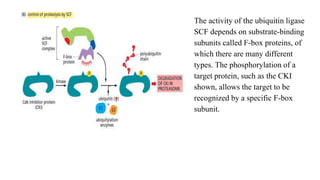
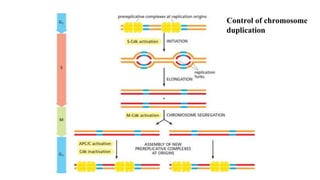
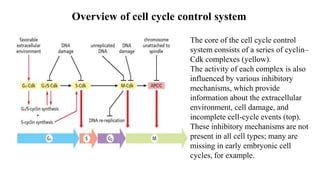
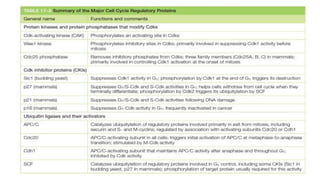
The cell cycle involves an orderly sequence of events where a cell duplicates its contents and divides into two daughter cells. It consists of interphase, where the cell grows and DNA replicates, and M phase where the cell divides. Key phases of interphase include G1, S, and G2 phases separated by gap phases. The cell cycle is tightly regulated by cyclins and CDKs which form complexes to drive the cell through checkpoints between phases. DNA replication only occurs once per cycle through control of initiation factors. Sister chromatids are held together by cohesin until anaphase.