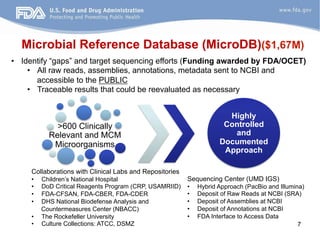
The document discusses the development of a regulatory-grade microbial reference database (MicroDB) by the FDA to support microbial identification and the detection of antimicrobial resistance through next-generation sequencing. It outlines objectives, requirements for genomic data, and collaborations with clinical labs to ensure high-quality sequencing data is accessible and traceable. The project emphasizes the need for robust, standardized microbial sequence databases to enhance clinical diagnostics and research efforts.

![2
Microbial NGS-Based Diagnostic Devices
• OIR/DMD working on a fast-tracked Draft Guidance
• On April 1st 2014 held Public Workshop
“Advancing Regulatory Science for High Throughput Sequencing
Devices for Microbial Identification and Detection of Antimicrobial
Resistance Markers” [FR Doc No: 2014-04940]
• Workshop agenda, discussion paper and webcast online:
http://www.fda.gov/MedicalDevices/NewsEvents/WorkshopsConferences/ucm386967.htm
Objectives:
1. Streamline/shorten clinical trials for microbial diagnosis/identification
2. Establish a new comparator algorithm for assays developed using this
new technology
3. Develop regulatory science standards for microbial genome sequencing
4. Investigate the regulatory science required for antimicrobial resistance
determination through microbial genome sequence information.](https://image.slidesharecdn.com/sichtightallonlmicrodbniststandards-141209162655-conversion-gate01/85/Development-of-FDA-MicroDB-A-Regulatory-Grade-Microbial-Reference-Database-2-320.jpg)