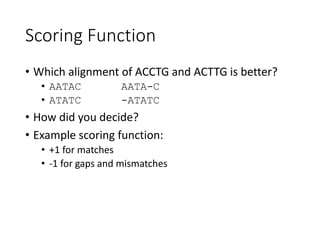
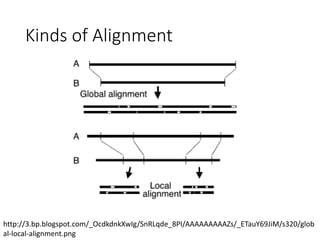
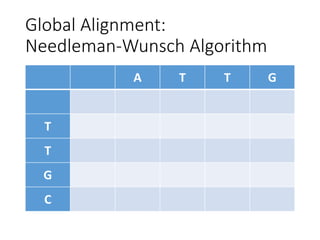
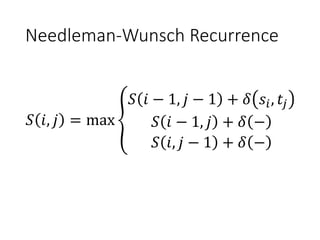
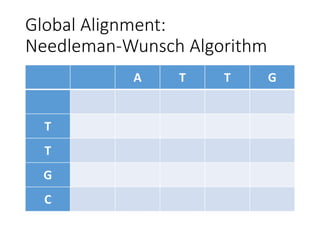
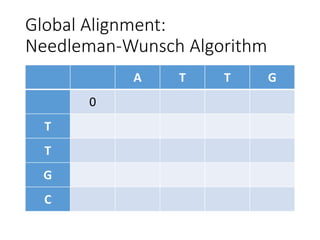
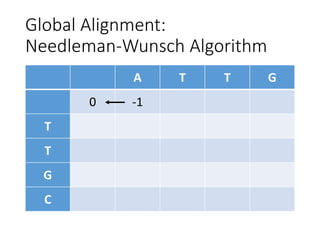
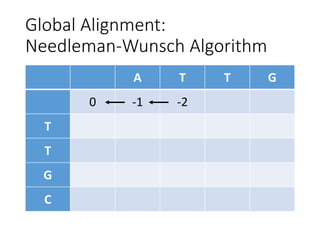
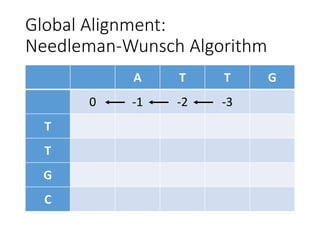
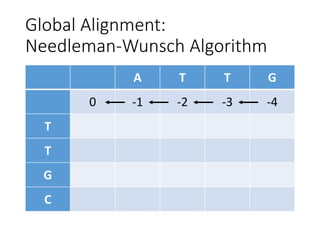
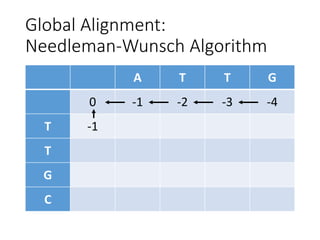
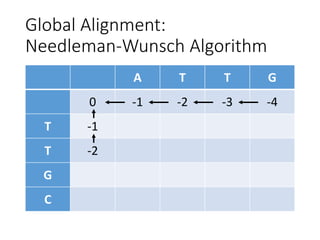
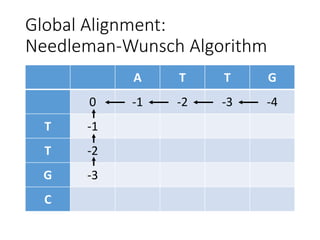
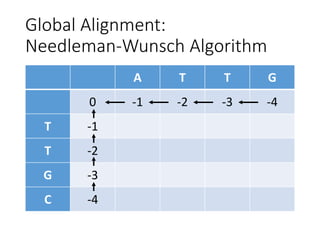
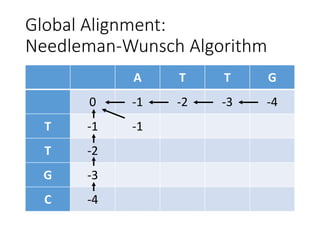
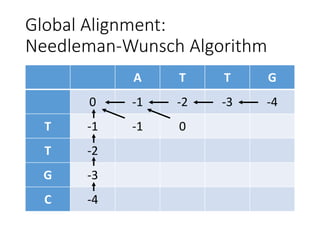
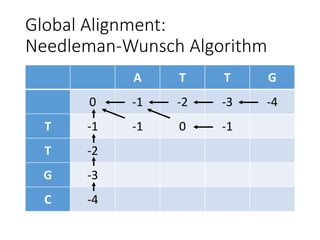
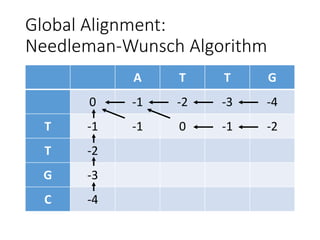
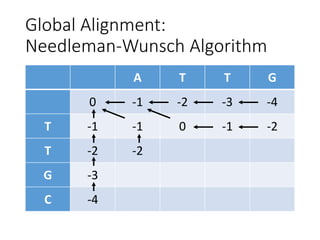
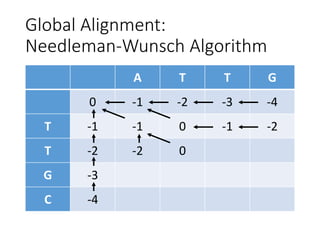
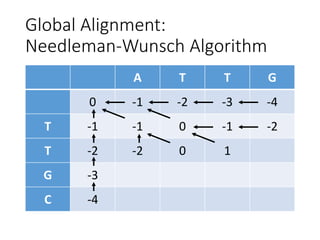
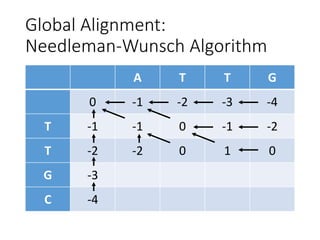
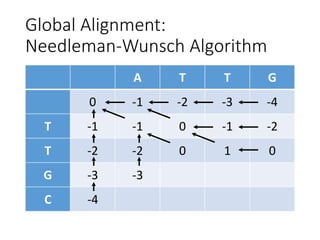
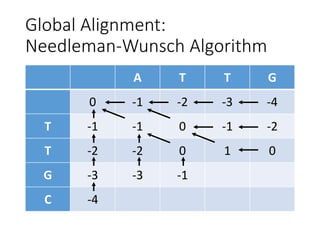
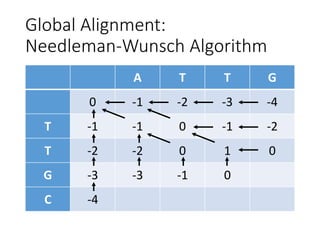
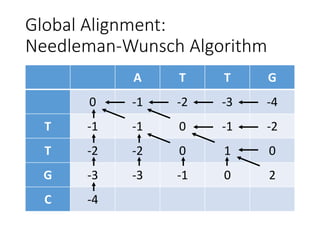
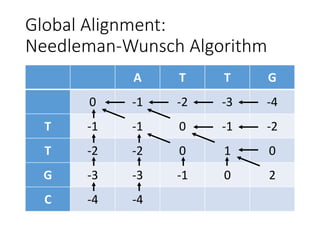
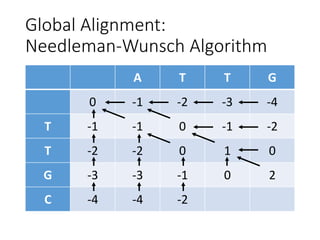
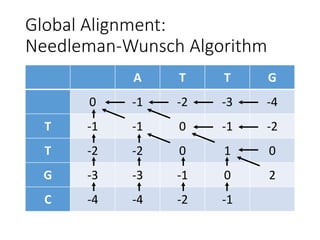
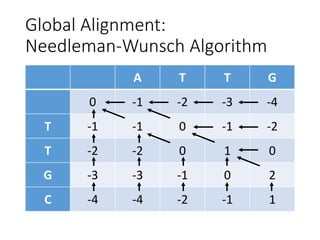
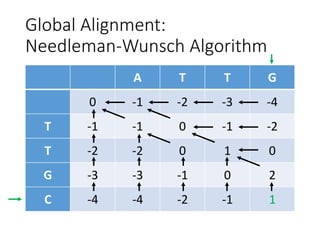
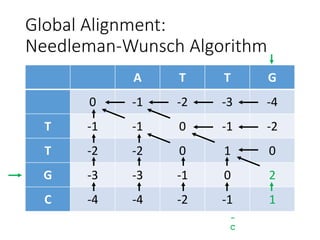
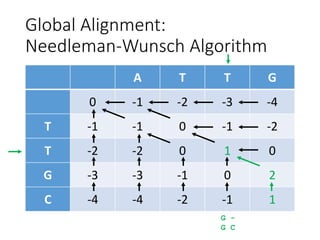
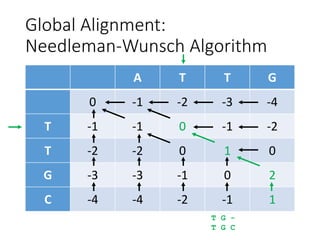
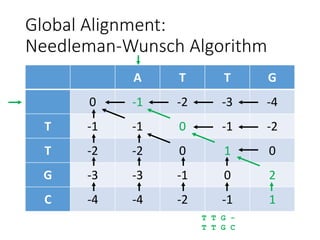
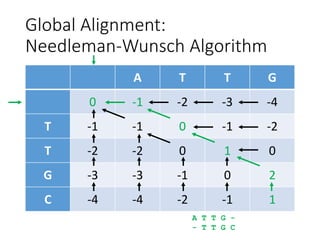
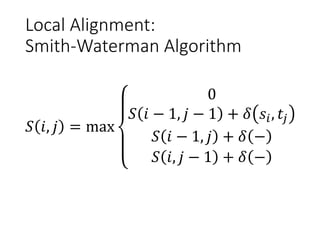
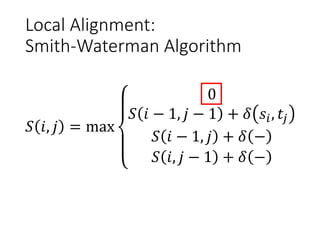
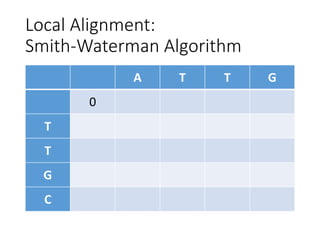
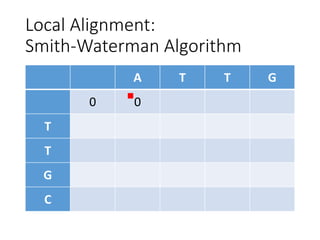
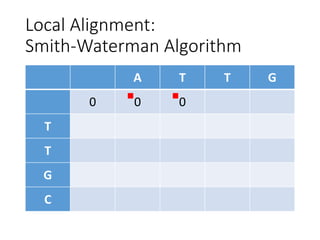
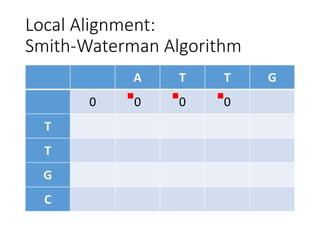
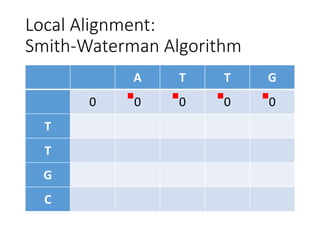
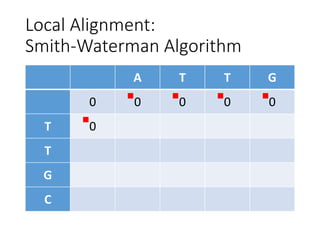
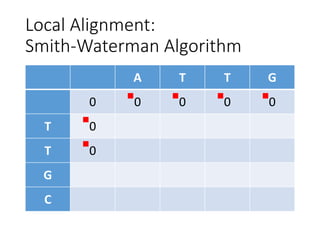
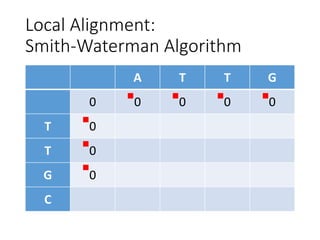
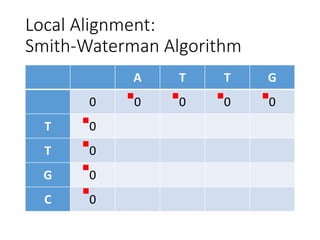
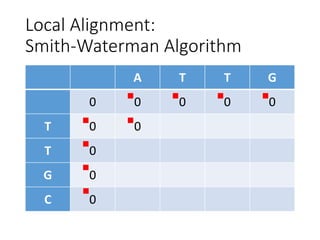
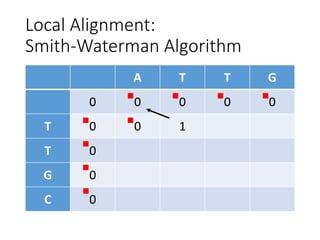
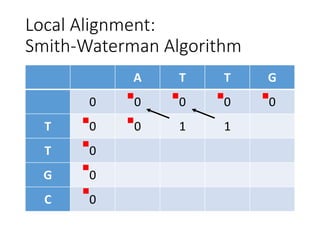
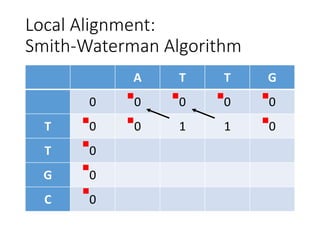
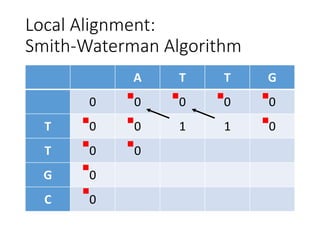
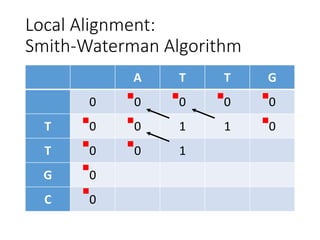
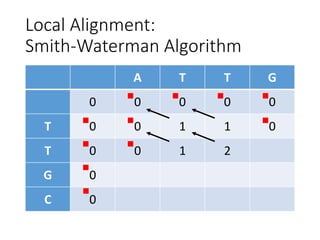
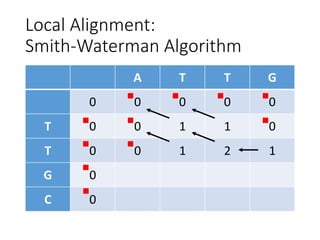
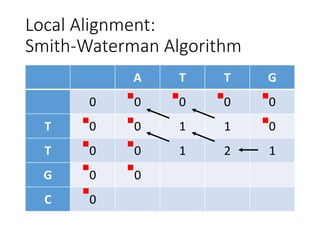
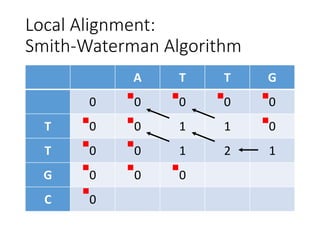
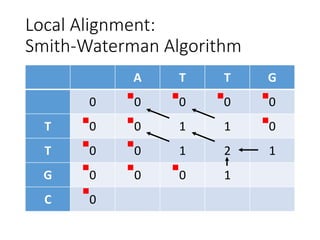
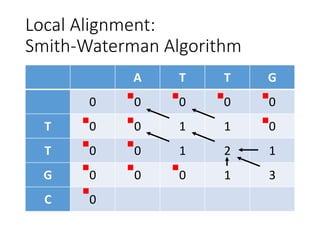
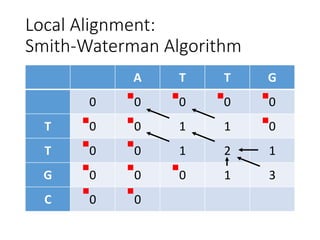
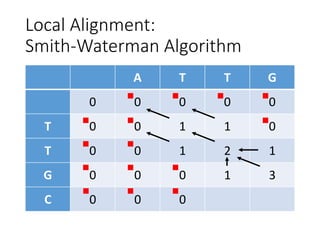
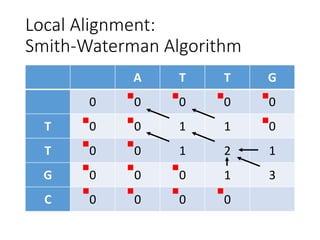
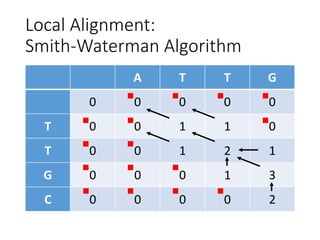
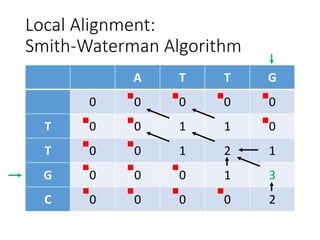
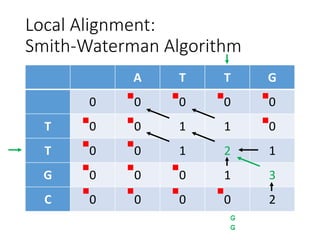
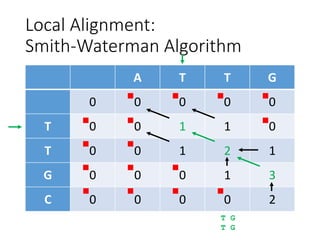
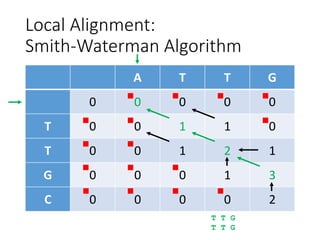
The document describes algorithms for sequence alignment, including the Needleman-Wunsch algorithm for global alignment and the Smith-Waterman algorithm for local alignment. It provides examples of filling in a scoring matrix according to the algorithms' recurrence relations and tracing back to find the optimal alignment between two sequences.