- The document summarizes Stanislav Andres' practical placement from July 1st to September 25th at the Latvian Institute of Organic Synthesis.
- The goals were to obtain and analyze the BBA03 protein involved in Lyme disease infection, and to perform routine analysis of biological samples using NMR and LCMS techniques.
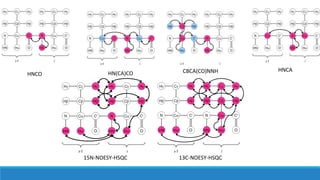
- NMR experiments were performed to assign the backbone and sidechain structures of the BBA03 protein, with the longest experiments taking 9 days to complete.