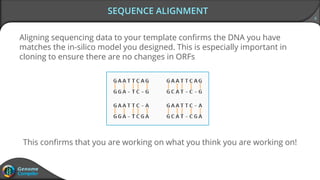
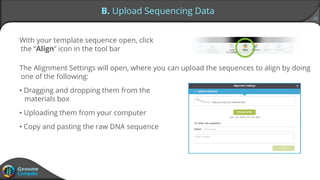
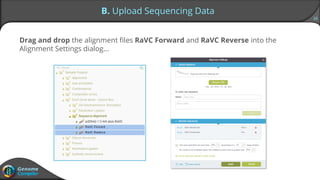
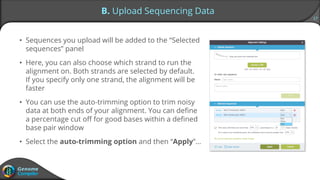
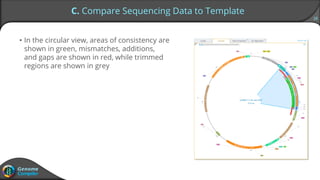
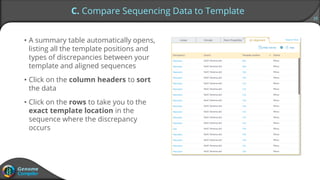
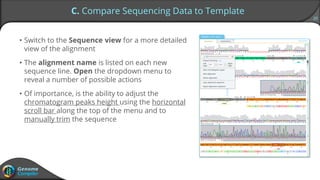
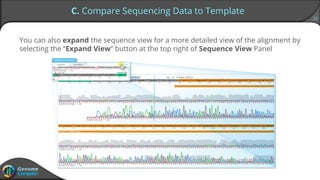
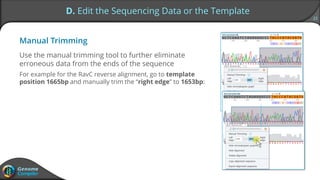
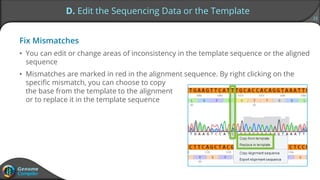
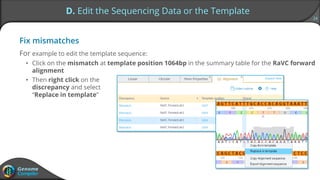
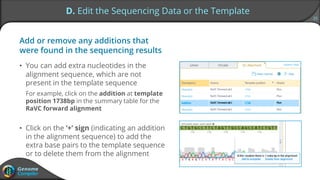
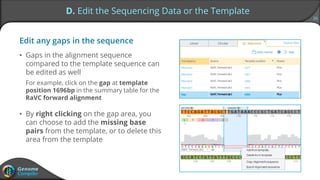
Sequence alignment can verify that cloned DNA matches the designed sequence by comparing sequencing data to a template. The pairwise alignment process identifies any discrepancies between the query sequence from sequencing data and the template. Editing functions allow fixing mismatches, additions, gaps, and ambiguous base calls by modifying either the sequencing data or template. After review and editing, the verified alignment can be copied or exported.