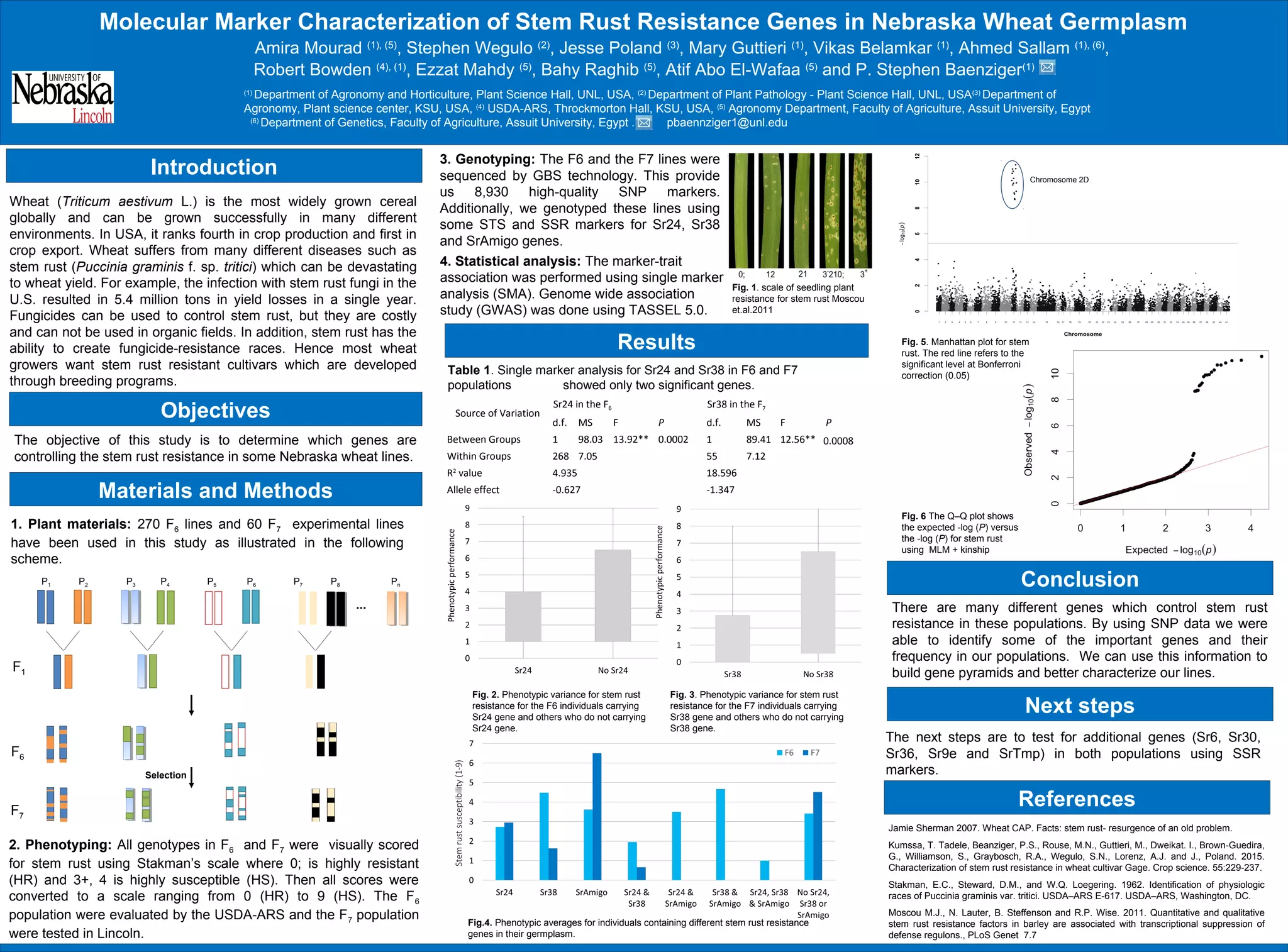
This study characterized stem rust resistance genes in Nebraska wheat germplasm. 270 F6 lines and 60 F7 lines were phenotyped for stem rust resistance and genotyped using GBS and marker analysis. Single marker analysis identified Sr24 as significantly associated with resistance in the F6 population and Sr38 in the F7 population. Genome-wide association also identified additional resistance genes. The study provides information on stem rust resistance genes in these wheat lines to enable gene pyramids for improved resistance breeding. Further testing will identify additional genes such as Sr6, Sr30, Sr36, Sr9e and SrTmp.