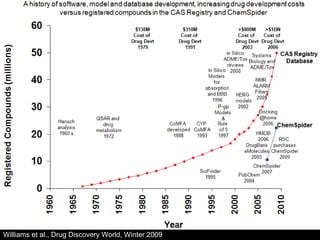
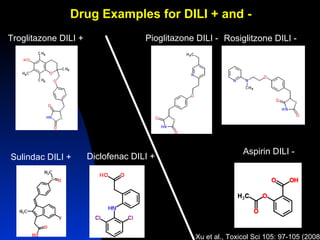
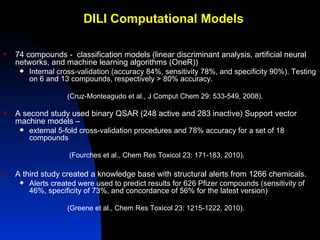
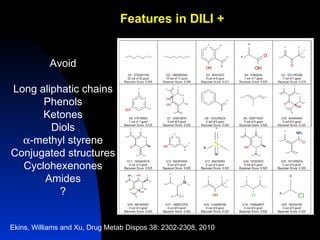
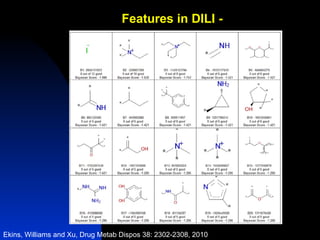
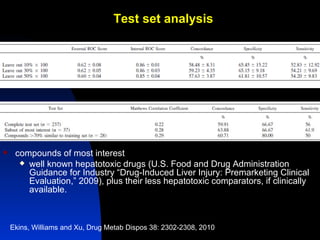
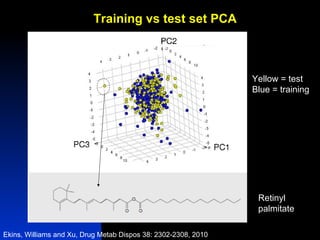
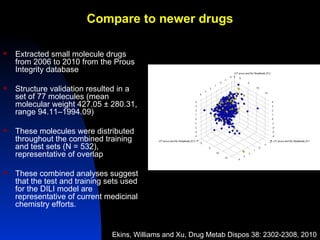
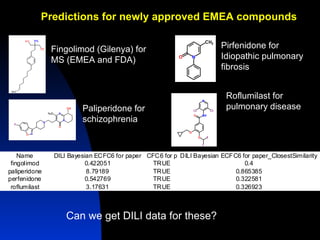
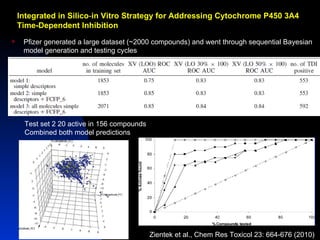
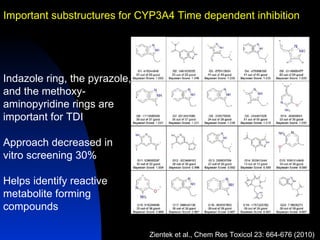
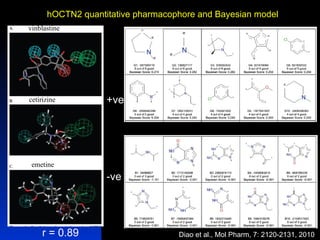
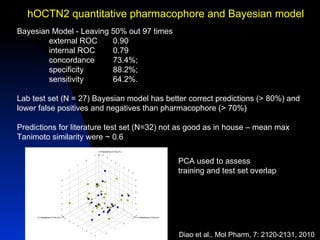
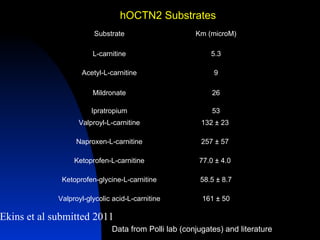
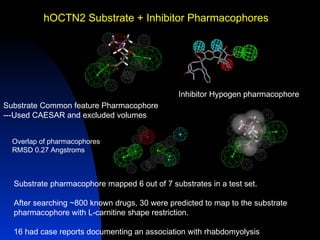
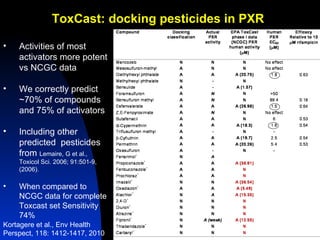
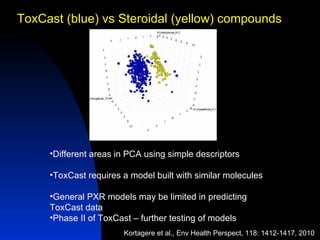
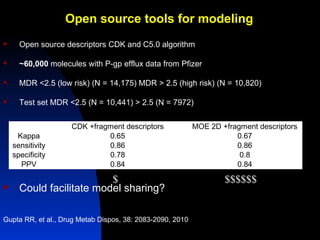
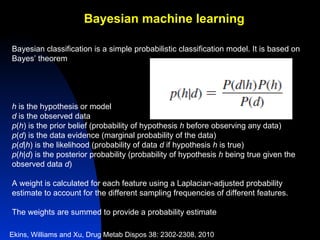
The document discusses computational models for predicting human toxicities, particularly focusing on drug-induced liver injury (DILI) and methodologies for assessing toxicity earlier in drug development. It highlights the use of various machine learning and classification models, including Bayesian classifiers, to improve the accuracy of predictions regarding drug safety. The document also emphasizes the importance of integrating in vitro and in silico approaches and the potential for shared models to enhance drug discovery efforts.