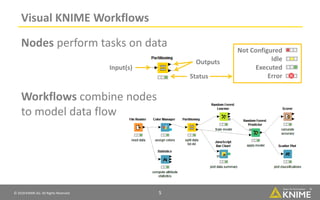
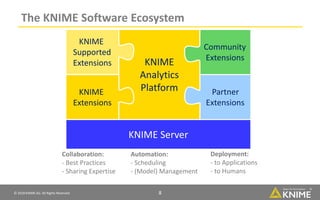
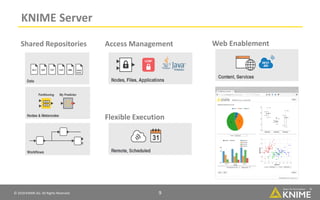
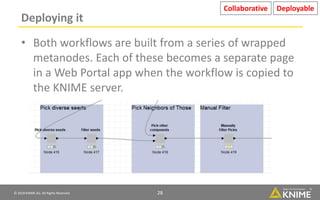
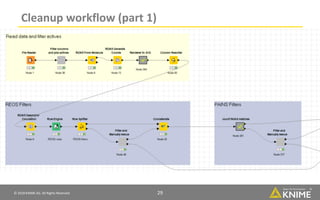
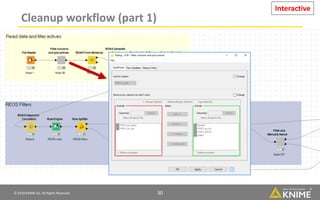
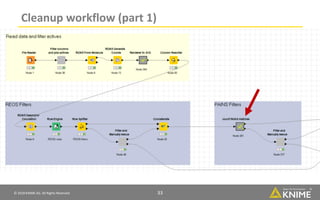
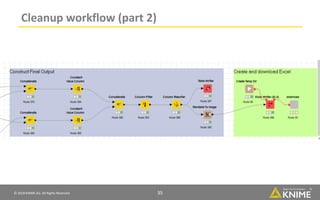
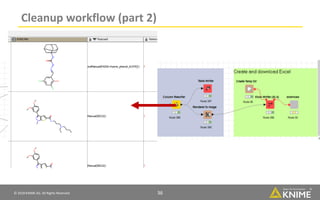
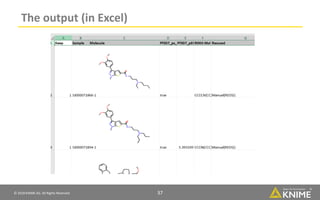
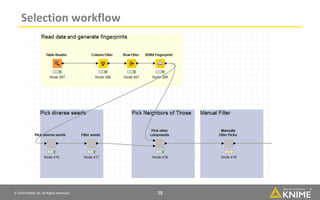
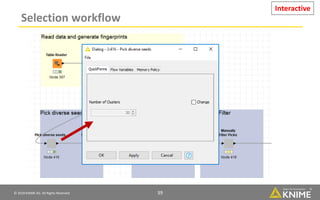
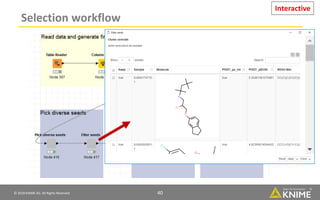
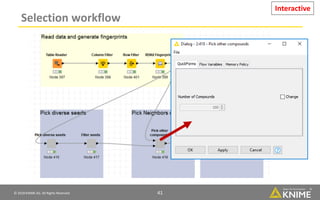
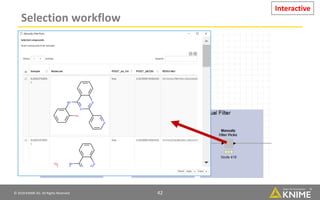
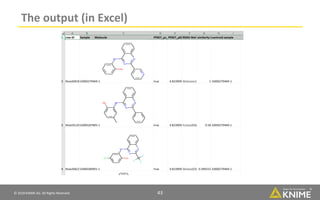
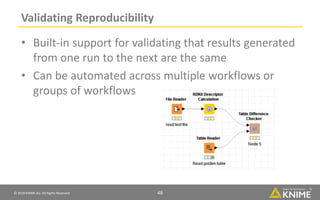
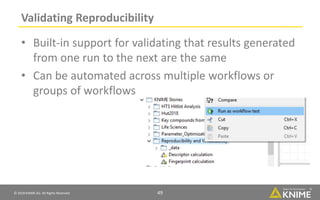
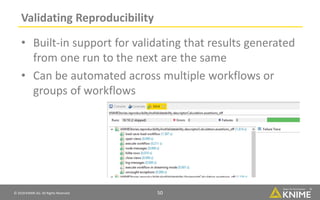
The document discusses a case study of using KNIME workflows to analyze a hit list from a high-throughput phenotypic screen for malaria in a reproducible and interactive manner. It describes using workflows to clean up the hit list by applying filters and selecting compounds for validation in a way that provides coverage of chemical space while also learning structure-activity relationships from the results. The workflows demonstrate how KNIME can help address common data analysis problems like repeatability, using multiple tools and data sources, and deploying and collaborating on analyses.