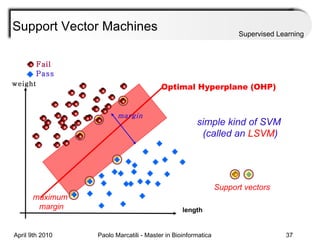
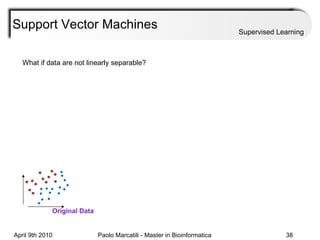
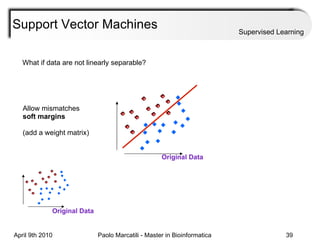
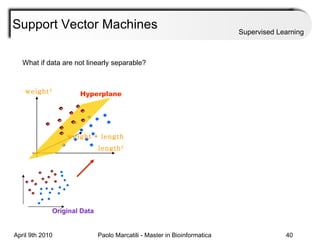
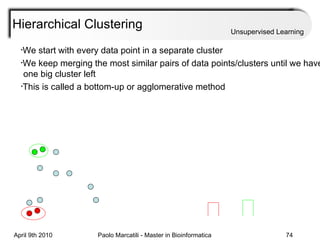
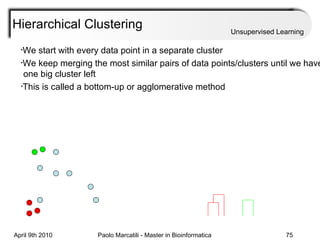
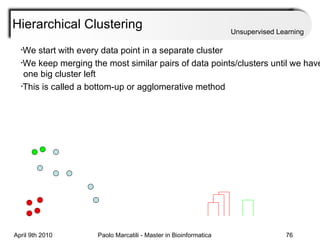
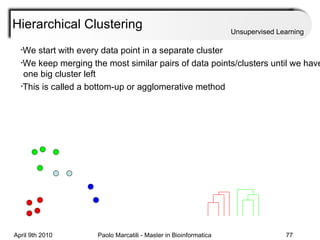
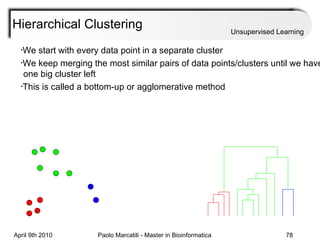
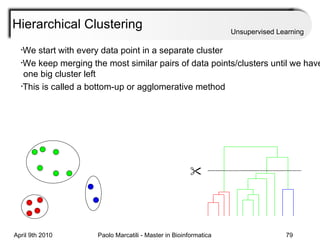
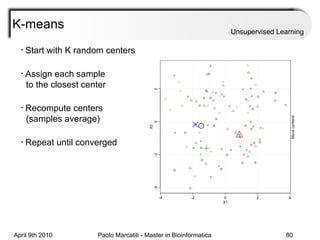
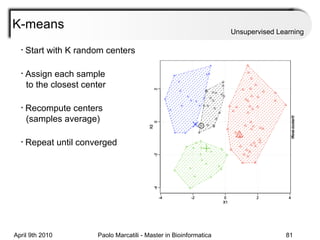
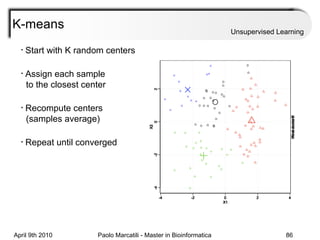
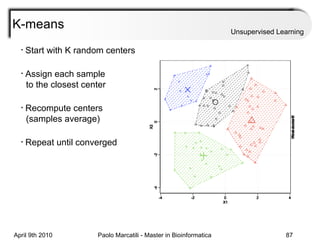
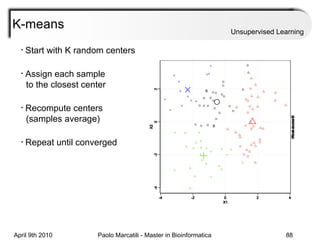
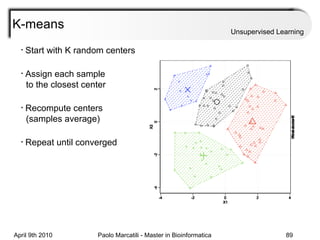
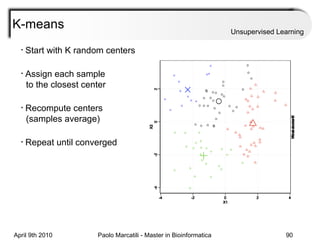
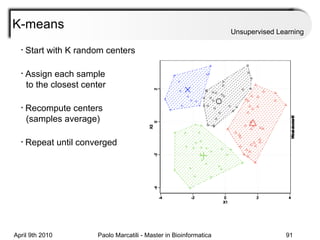
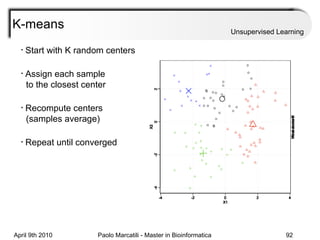
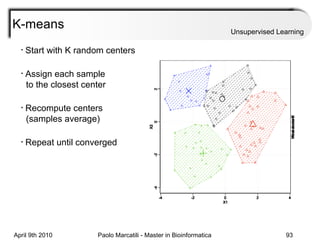
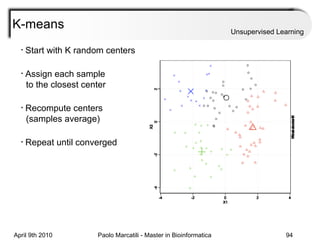
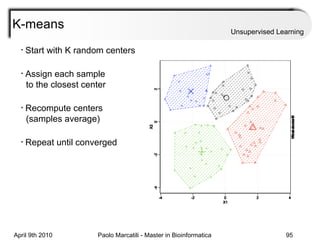
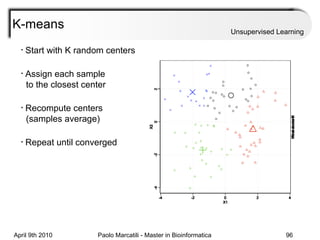
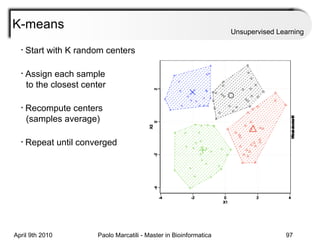
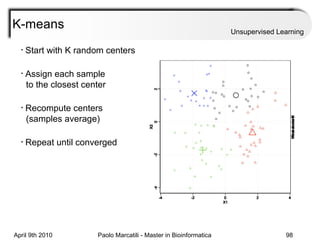
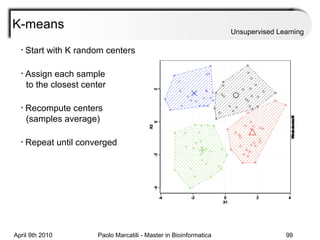
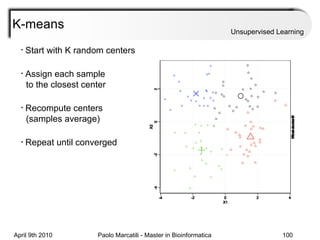
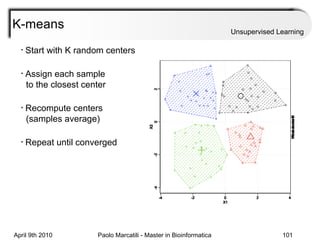
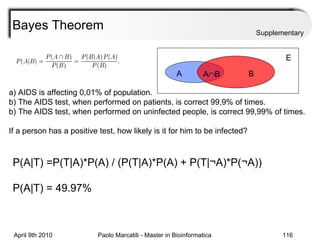
This document provides an overview of machine learning methods, including supervised and unsupervised learning. It discusses commonly used machine learning algorithms like support vector machines (SVM), hidden Markov models, decision trees, random forests, Bayesian networks, and neural networks. It also covers datasets, assessment metrics, and caveats to consider when using machine learning.
![Machine Learning Methods: an overview Master in Bioinformatica – April 9th, 2010 Paolo Marcatili University of Rome “Sapienza” Dept. of Biochemical Sciences “Rossi Fanelli” [email_address] Overview Supervised Learning Unsupervised Learning Caveats](https://image.slidesharecdn.com/machine-learning-arial-1318413710-phpapp01-111012050429-phpapp01/75/Machine-Learning-1-2048.jpg)





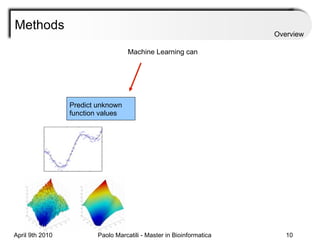
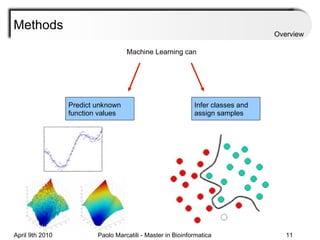
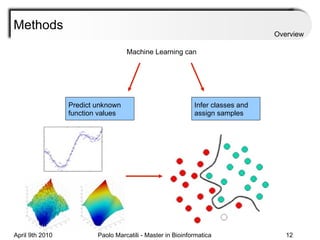
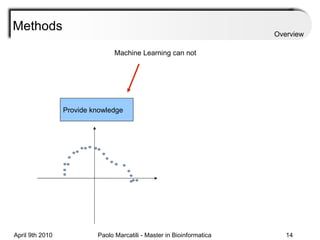
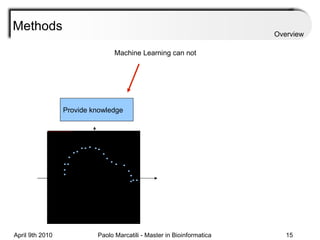
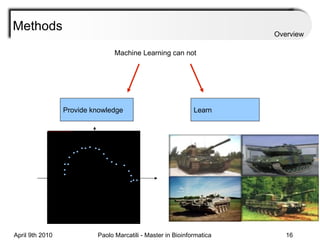
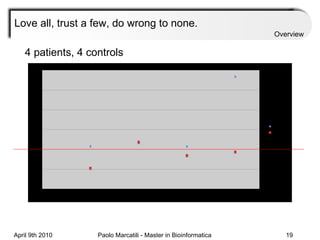
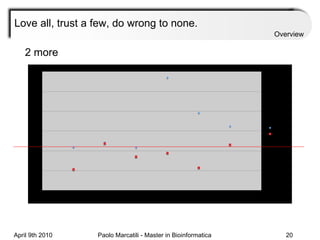
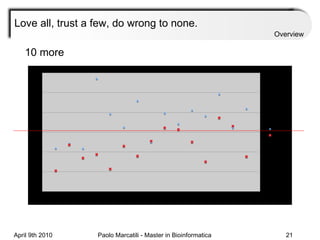
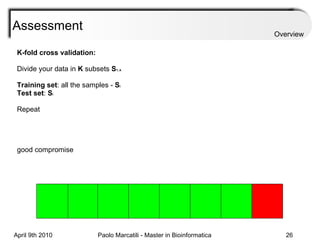
![Assessment Sensitivity: TP/ [ TP + FN ] Given the disease is present, the likelihood of testing positive. Specificity: TN / [ TN + FP ] Given the disease is not present, the likelihood of testing negative. Positive Predictive Value : TP / [ TP + FP ] Given a test is positive, the likelihood disease is present Overview](https://image.slidesharecdn.com/machine-learning-arial-1318413710-phpapp01-111012050429-phpapp01/85/Machine-Learning-27-320.jpg)
![Assessment Sensitivity: TP/ [ TP + FN ] Given the disease is present, the likelihood of testing positive. Specificity: TN / [ TN + FP ] Given the disease is not present, the likelihood of testing negative. Positive Predictive Value : TP / [ TP + FP ] Given a test is positive, the likelihood disease is present receiver operating characteristic (ROC) is a graphical plot of the sensitivity vs. (1 - specificity) for a binary classifier system as its discrimination threshold is varied. Overview](https://image.slidesharecdn.com/machine-learning-arial-1318413710-phpapp01-111012050429-phpapp01/85/Machine-Learning-28-320.jpg)
![Assessment Overview Sensitivity: TP/ [ TP + FN ] Given the disease is present, the likelihood of testing positive. Specificity: TN / [ TN + FP ] Given the disease is not present, the likelihood of testing negative. Predictive Value Positive: TP / [ TP + FP ] Given a test is positive, the likelihood disease is present receiver operating characteristic (ROC) is a graphical plot of the sensitivity vs. (1 - specificity) for a binary classifier system as its discrimination threshold is varied. Area under ROC (AROC) is often used as a parameter to compare different classifiers](https://image.slidesharecdn.com/machine-learning-arial-1318413710-phpapp01-111012050429-phpapp01/85/Machine-Learning-29-320.jpg)