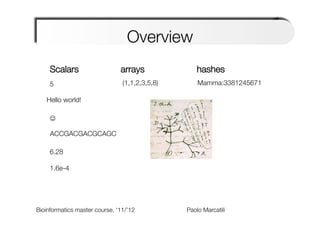
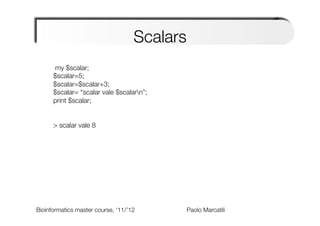
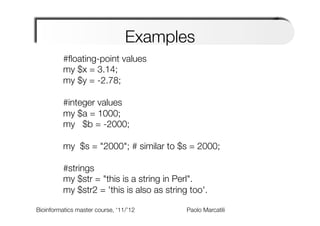
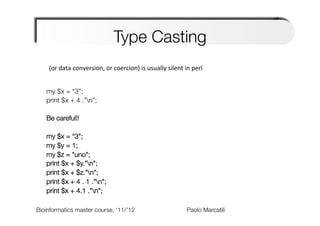
The document provides an overview and introduction to data types in Perl, including scalars, arrays, and hashes. Scalars can hold single data values like numbers or strings. Arrays hold ordered collections of scalars and are indexed with square brackets. Hashes store key-value pairs and are indexed with curly brackets. The document discusses how to define, access, and manipulate each of these data types in Perl code examples. It also covers type casting, operations, and common functions for each type like sorting arrays and hashes.














![array - 2
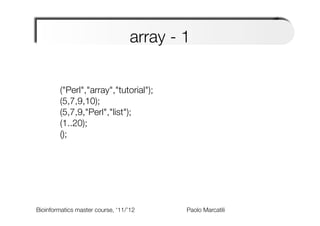
my @str_array=("Perl","array","tutorial");
my @num_array=(5,7,9,10);
my @mixed_array=(5,7,9,"Perl","list");
my @rg_array=(1..20);
my @empty_array=();
print $str_array[1]; # 1st element is [0]
Bioinformatics master course, ‘11/’12
Paolo Marcatili](https://image.slidesharecdn.com/masterdatatypes2011-120113063355-phpapp02/85/Master-datatypes-2011-25-320.jpg)



![Hashes
• Hashes are like array, they store collections of scalars"
... but unlike arrays, indexing is by name (just like in
real life!!!)"
• Two components to each hash entry:
– Key
example : name
– Value
example : phone number
• Hashes denoted with %
– Example : %phoneDirectory
• Elements are accessed using {} (like [] in arrays)
Bioinformatics master course, ‘11/’12
Paolo Marcatili](https://image.slidesharecdn.com/masterdatatypes2011-120113063355-phpapp02/85/Master-datatypes-2011-29-320.jpg)