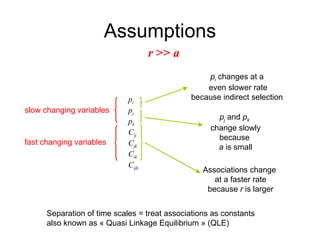
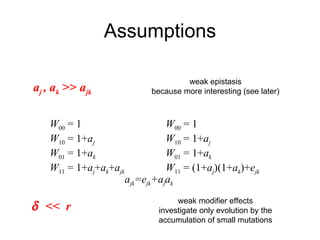
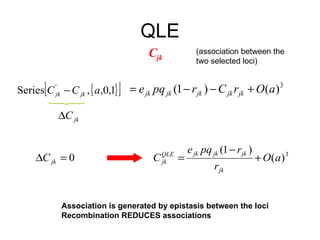
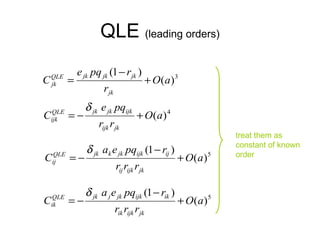
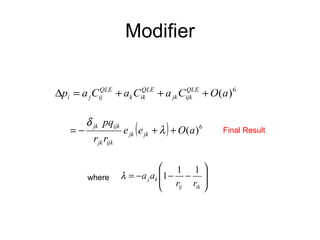
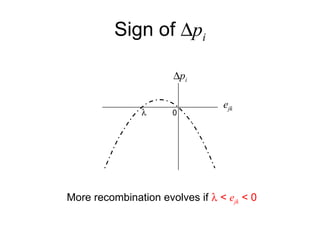
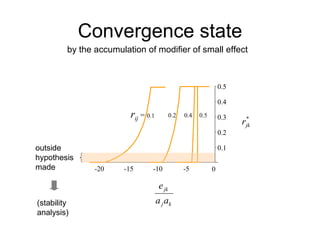
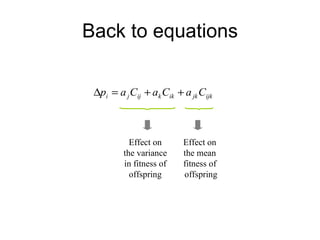
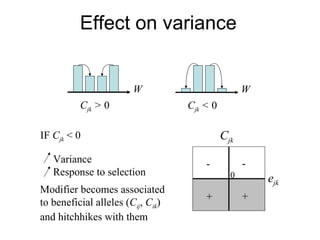
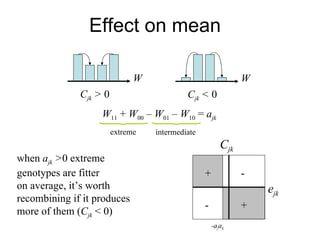
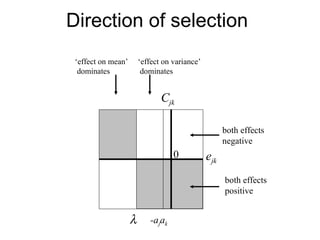
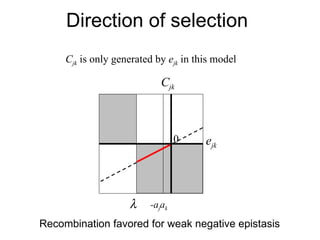
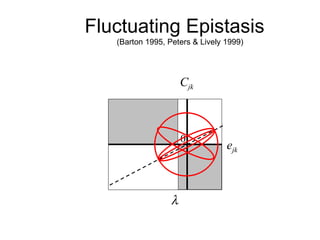
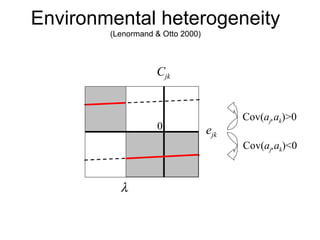
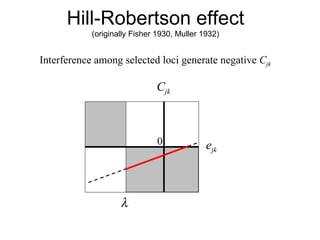
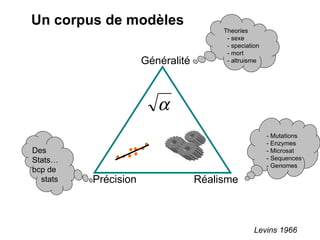
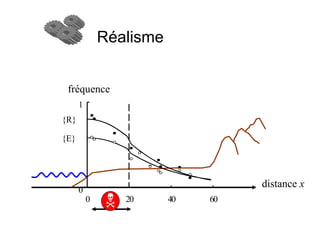
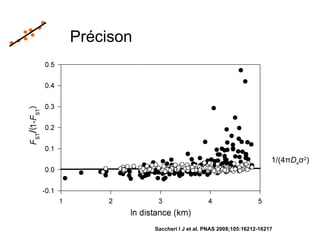
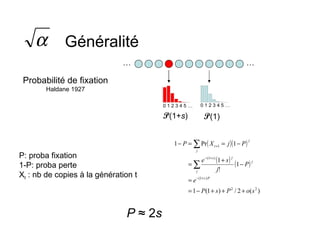
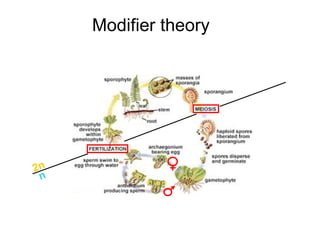
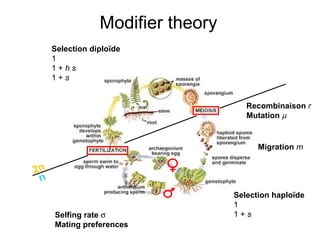
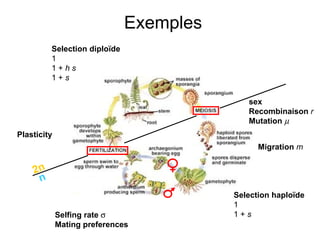
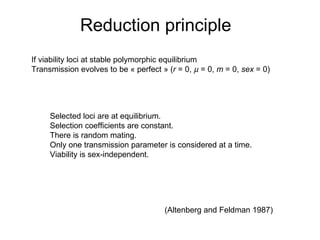
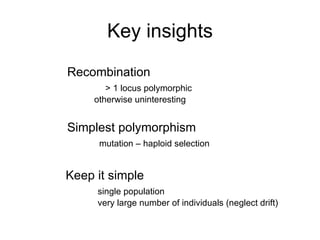
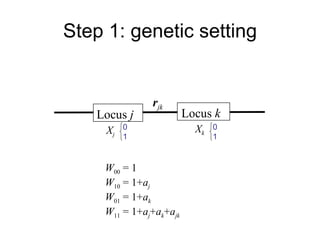
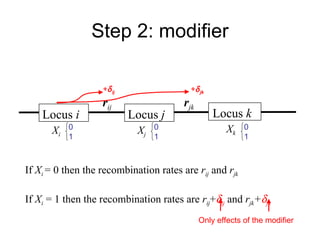
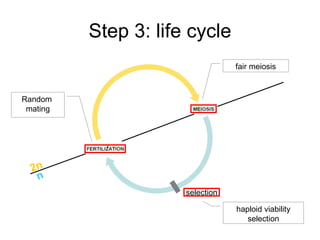
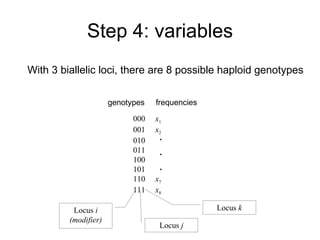
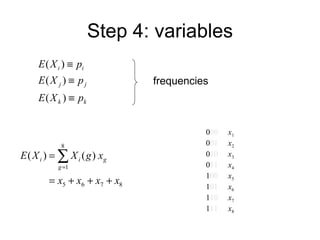
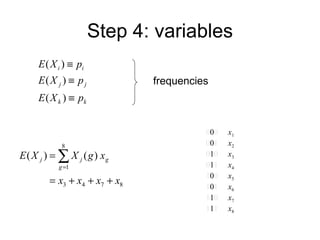
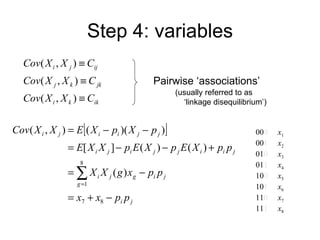
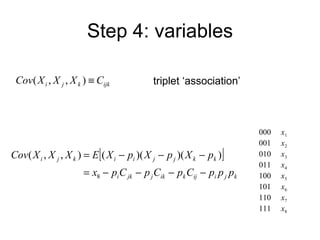
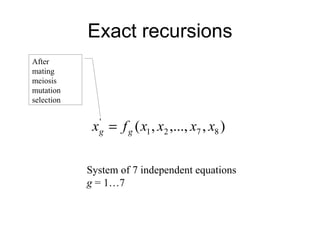
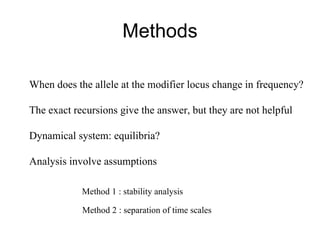
This document summarizes a population genetics model for studying the evolution of recombination rates. The model considers three loci, including a modifier locus that can alter the recombination rates between two selected loci. Using recursion equations and assumptions like weak epistasis and separation of timescales, the model analyzes when and why the frequency of a recombination-increasing allele at the modifier locus may increase over time through indirect selection. The key results are that more recombination evolves if epistasis between the selected loci is weakly negative and their association is negative.







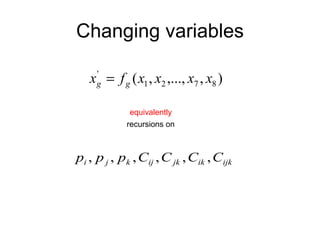

![Taylor Series constant approximation linear approximation quadratic approximation Notation : Series[ f ( x ),{ x , x 0 , 2}] (like in Mathematica)](https://image.slidesharecdn.com/lenormandgenetpop-100610074937-phpapp02/85/Thomas-Lenormand-Genetique-des-populations-39-320.jpg)