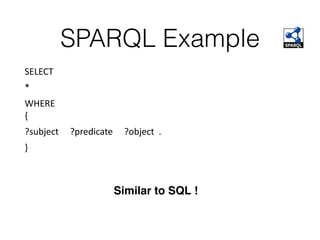
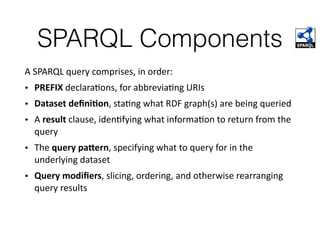
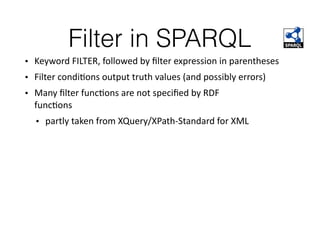
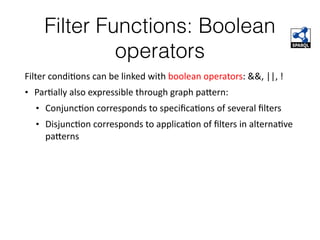
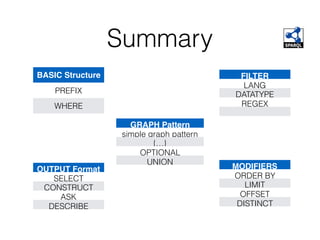
This document discusses SPARQL, the SPARQL Protocol and RDF Query Language. SPARQL is used to query and retrieve data stored in RDF format. The key components of a SPARQL query are PREFIX declarations, a dataset definition, a result clause specifying what to return, a query pattern matching the dataset, and optional query modifiers. SPARQL supports operations like filtering results, sorting, limiting output and removing duplicates. Examples show how to query RDF data from public SPARQL endpoints like DBpedia using these features.