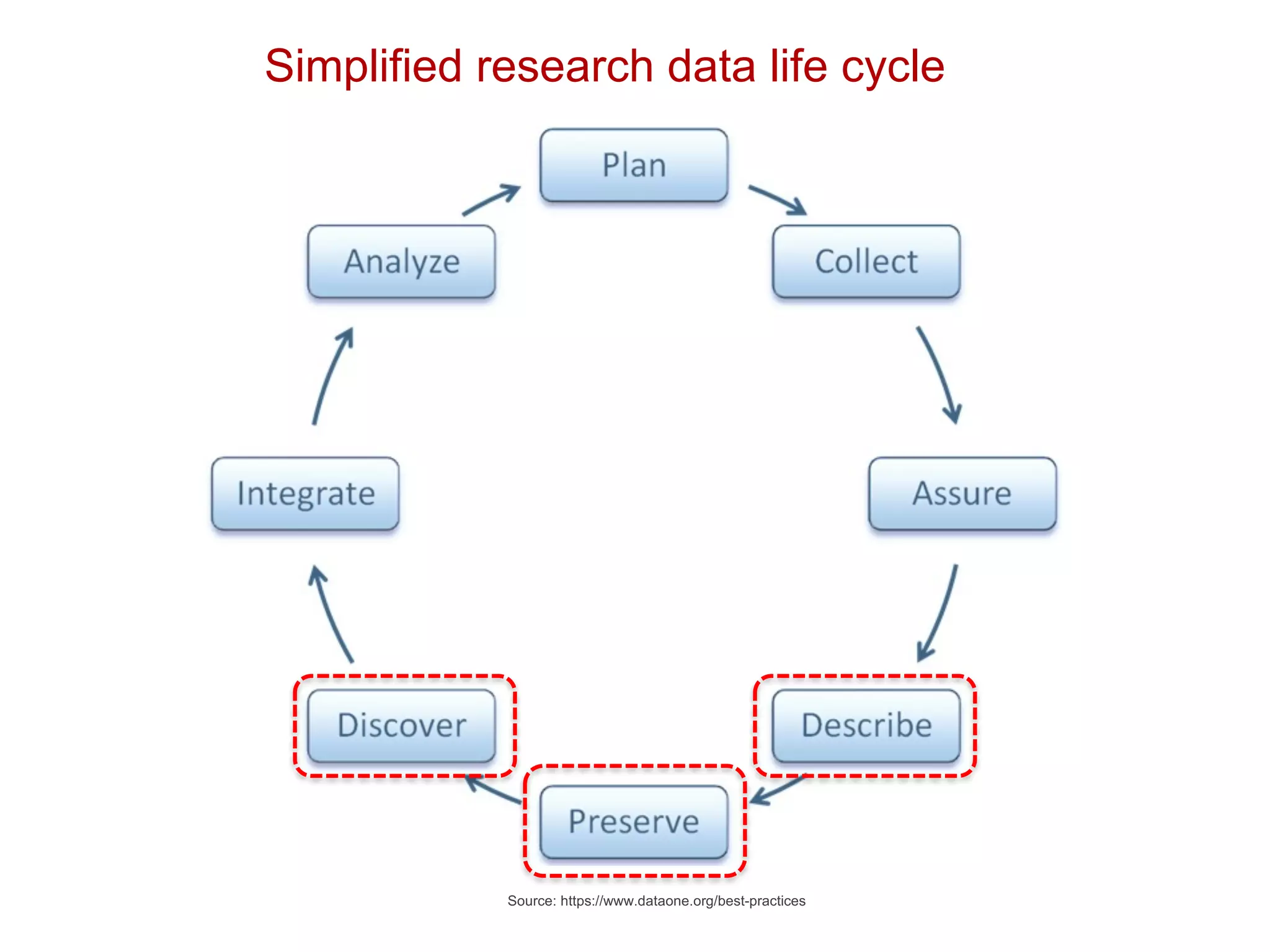
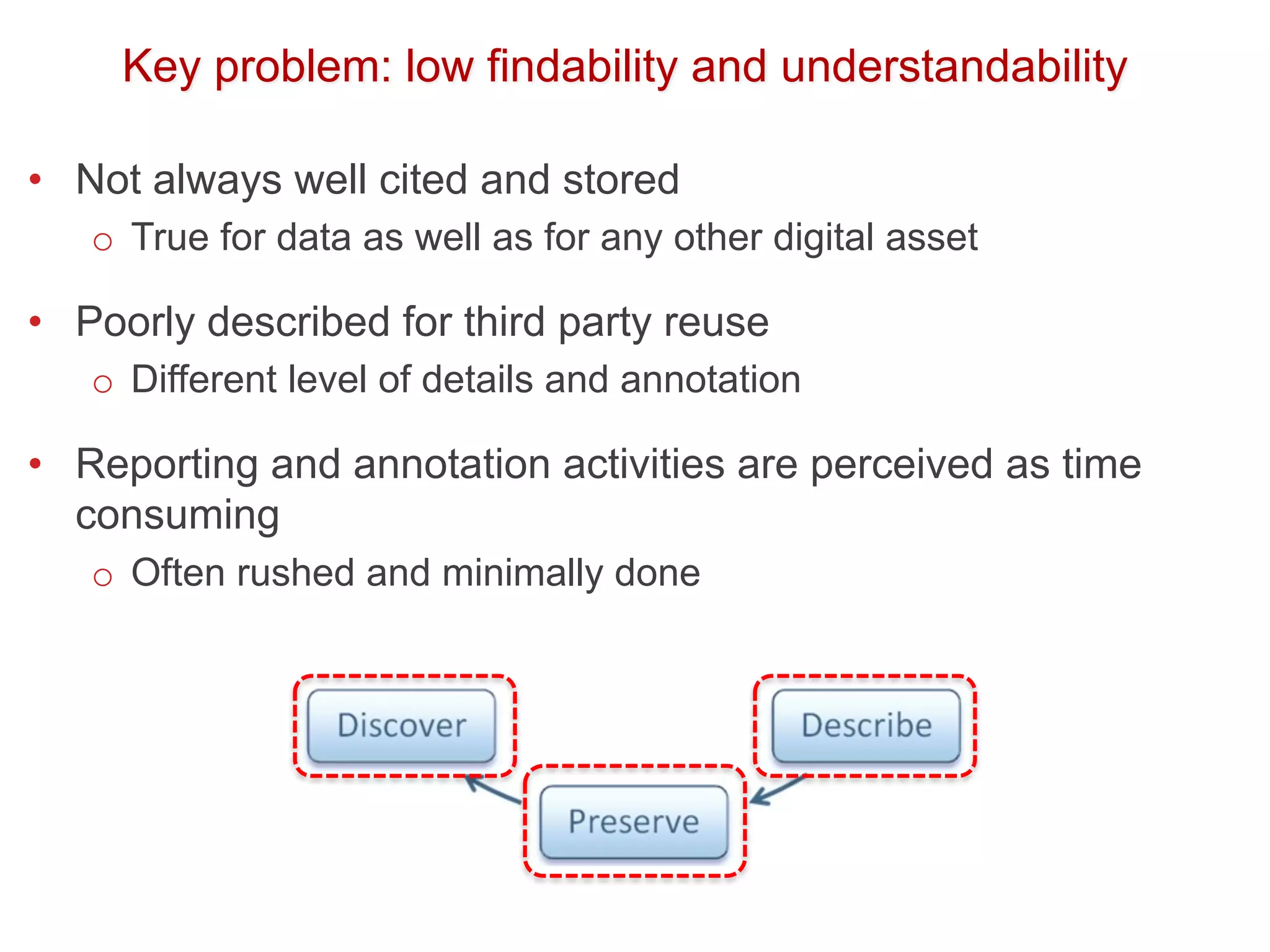
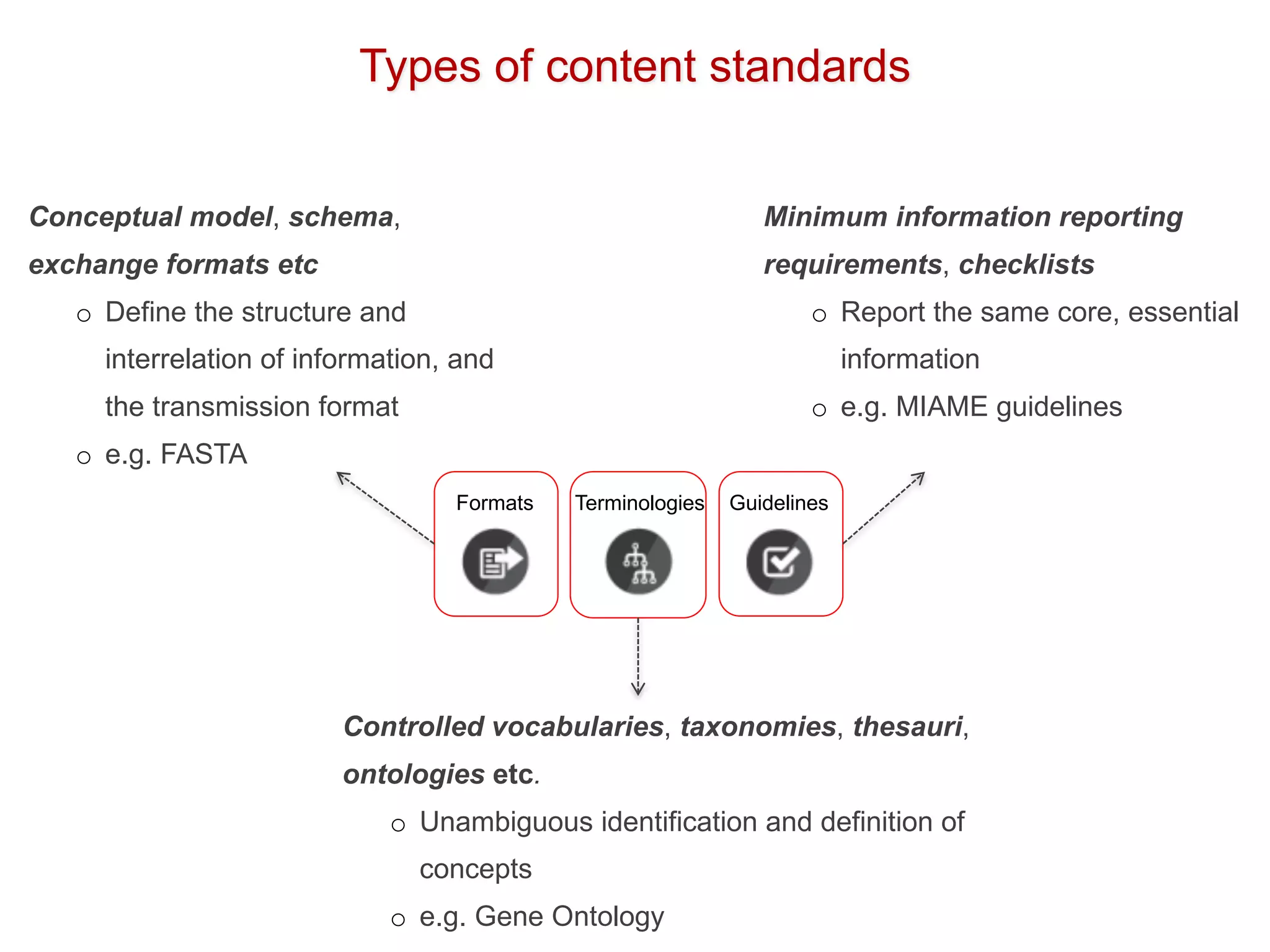
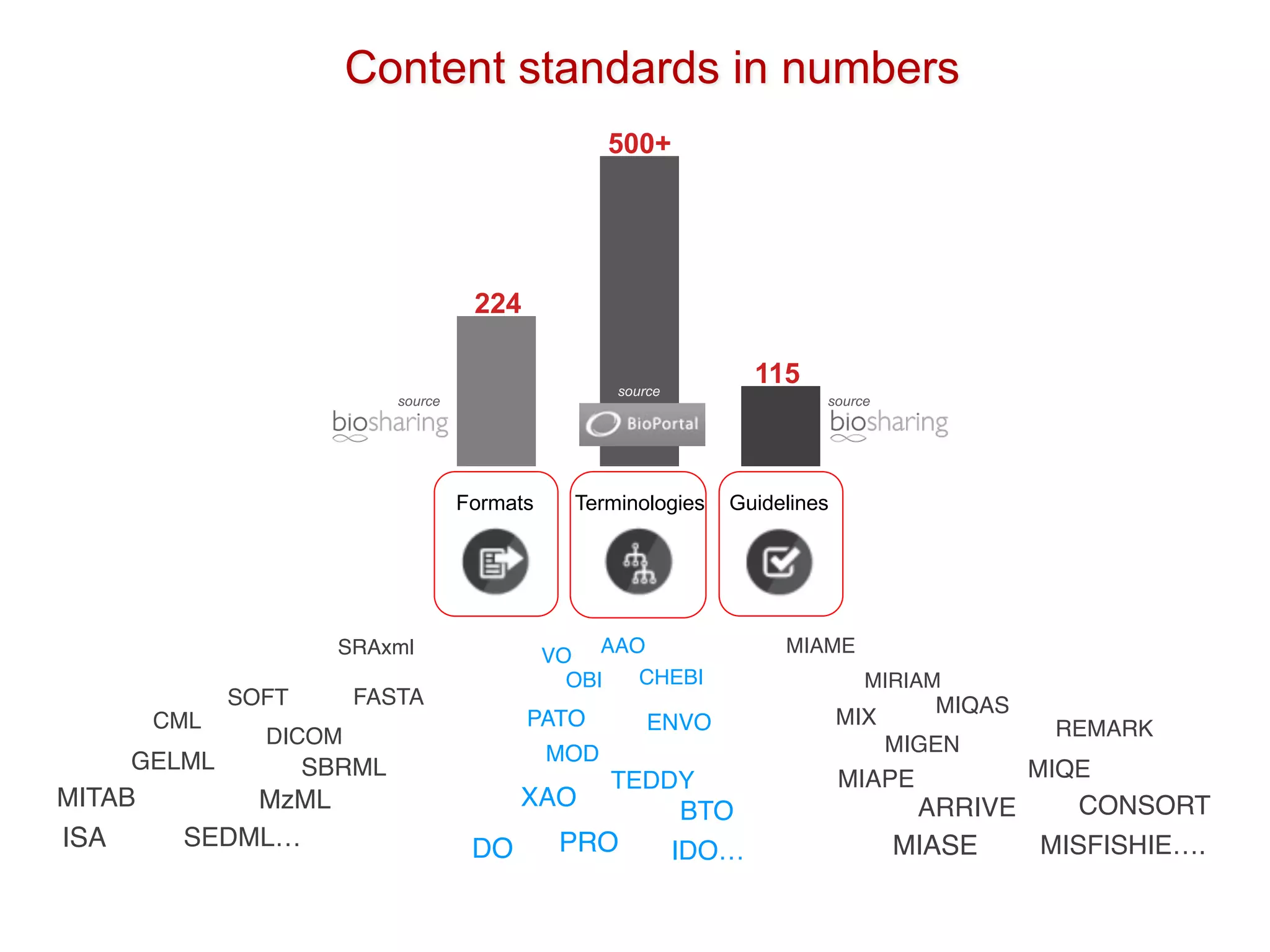
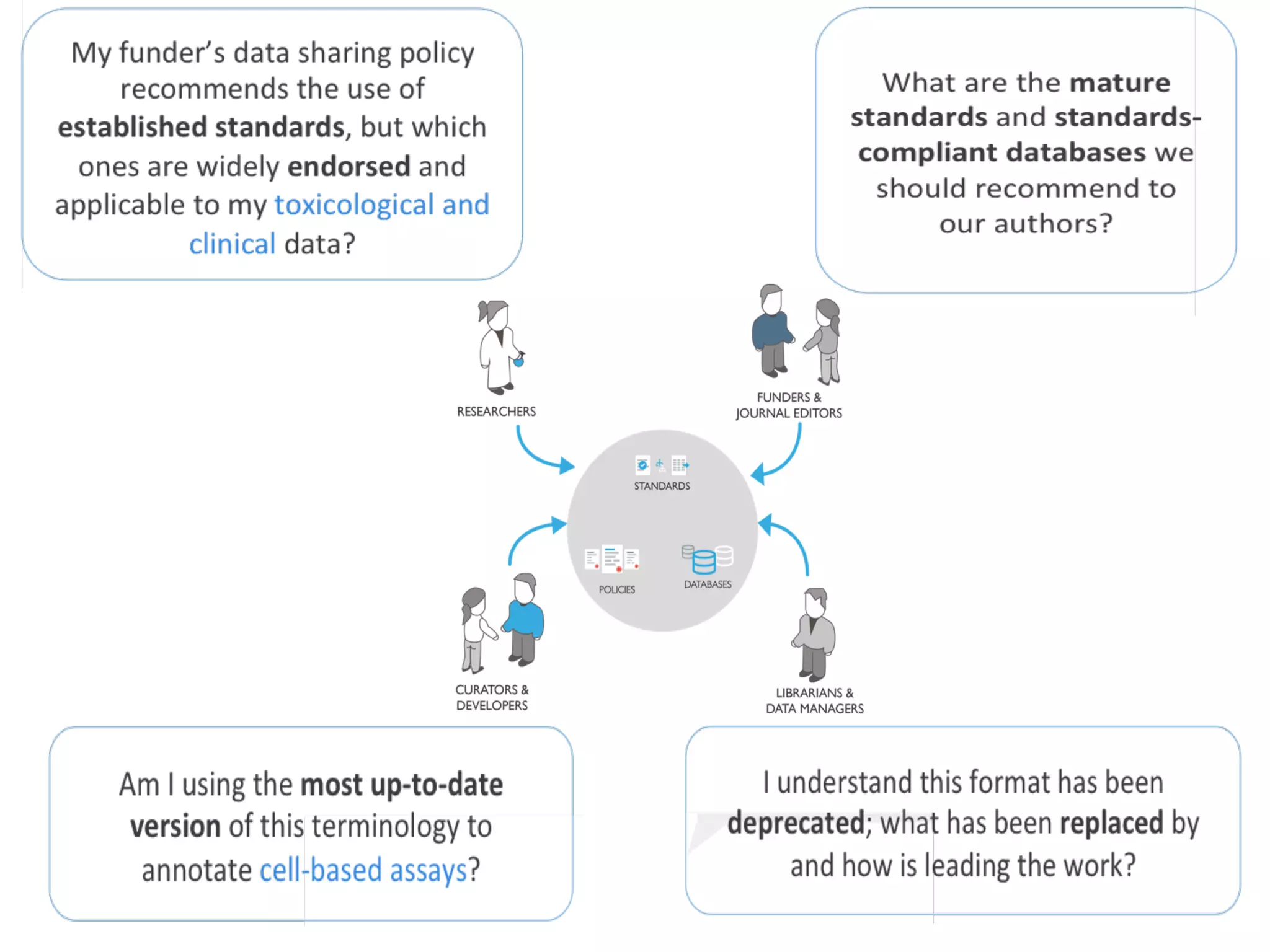
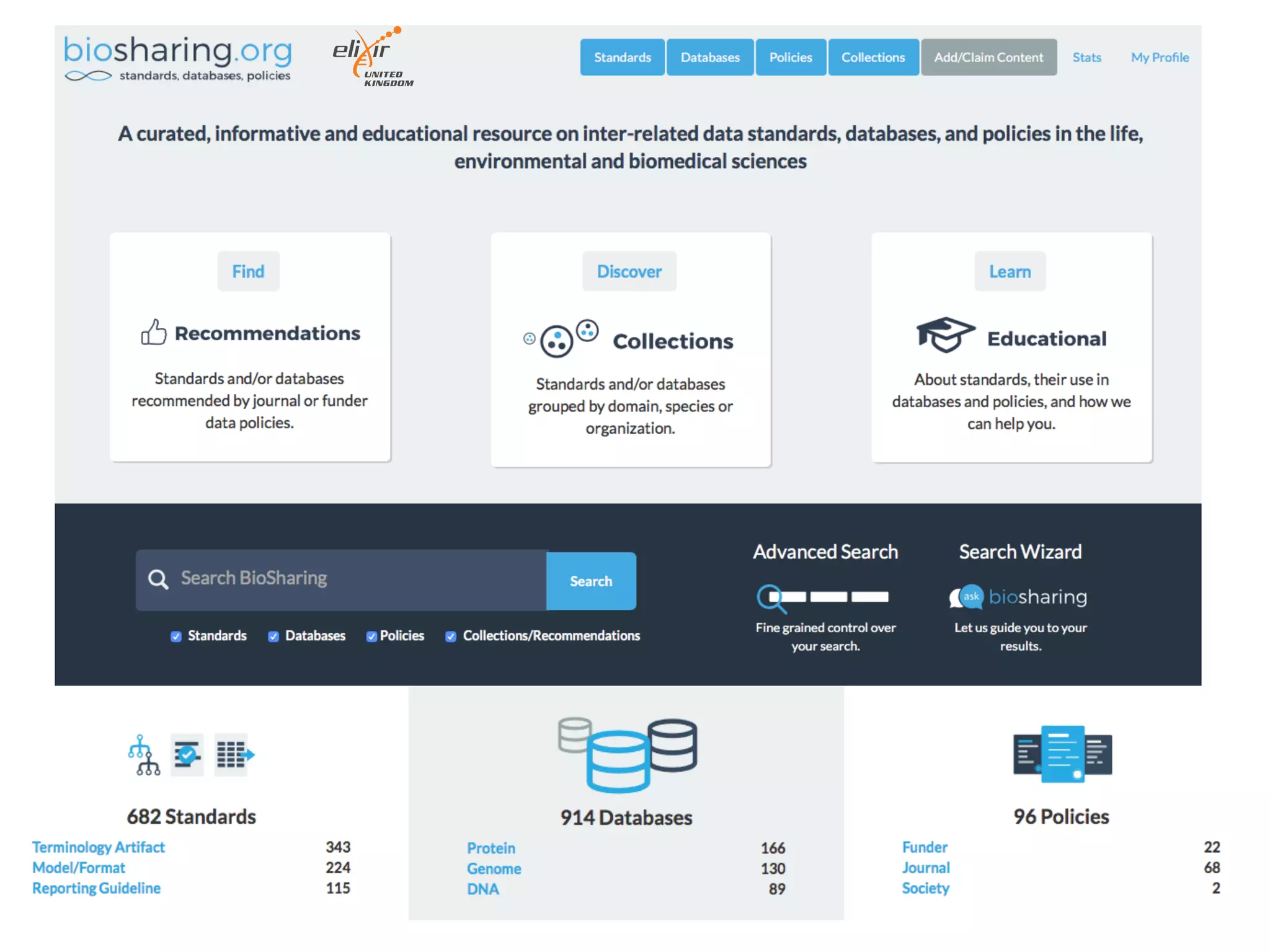
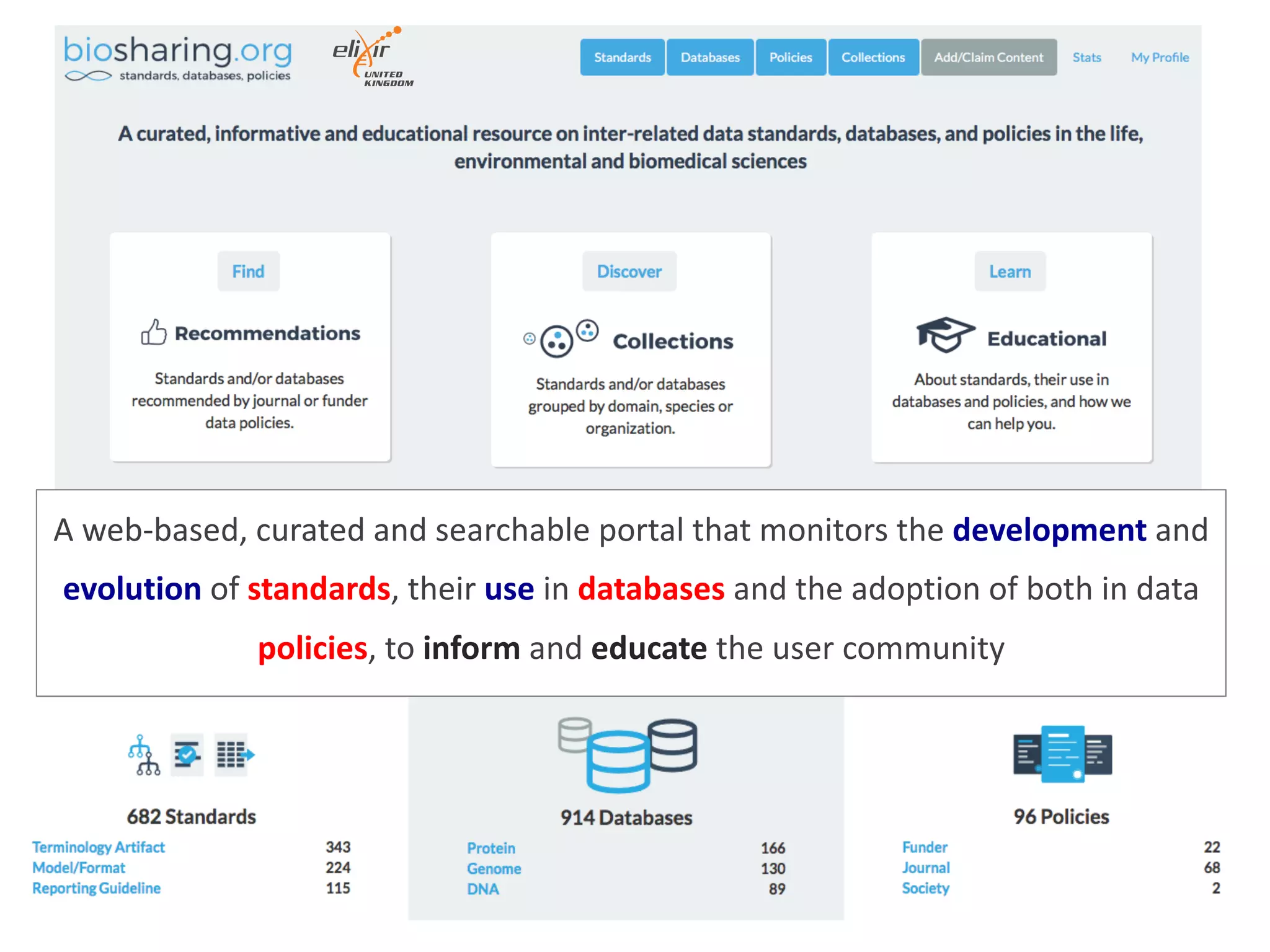
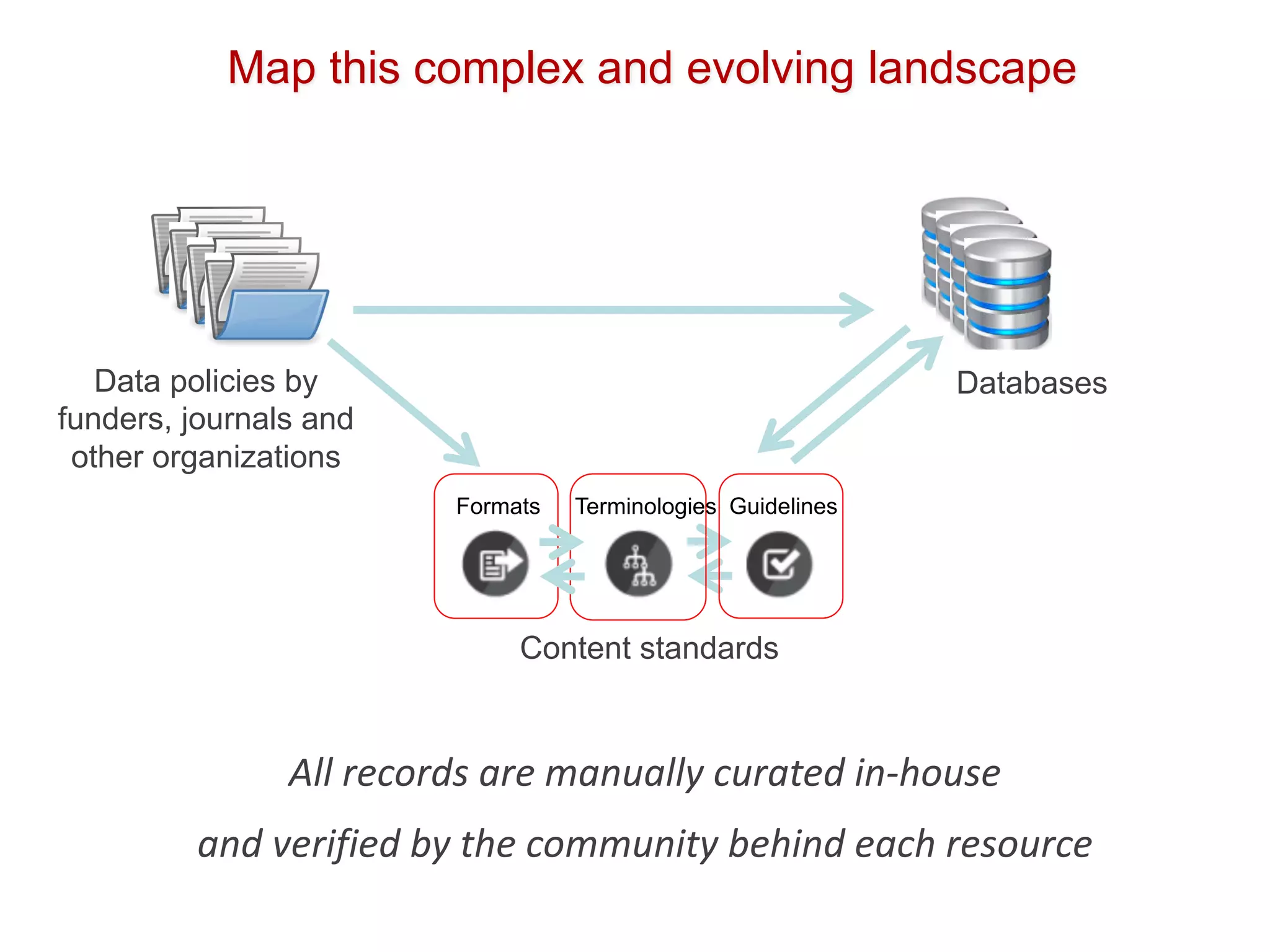
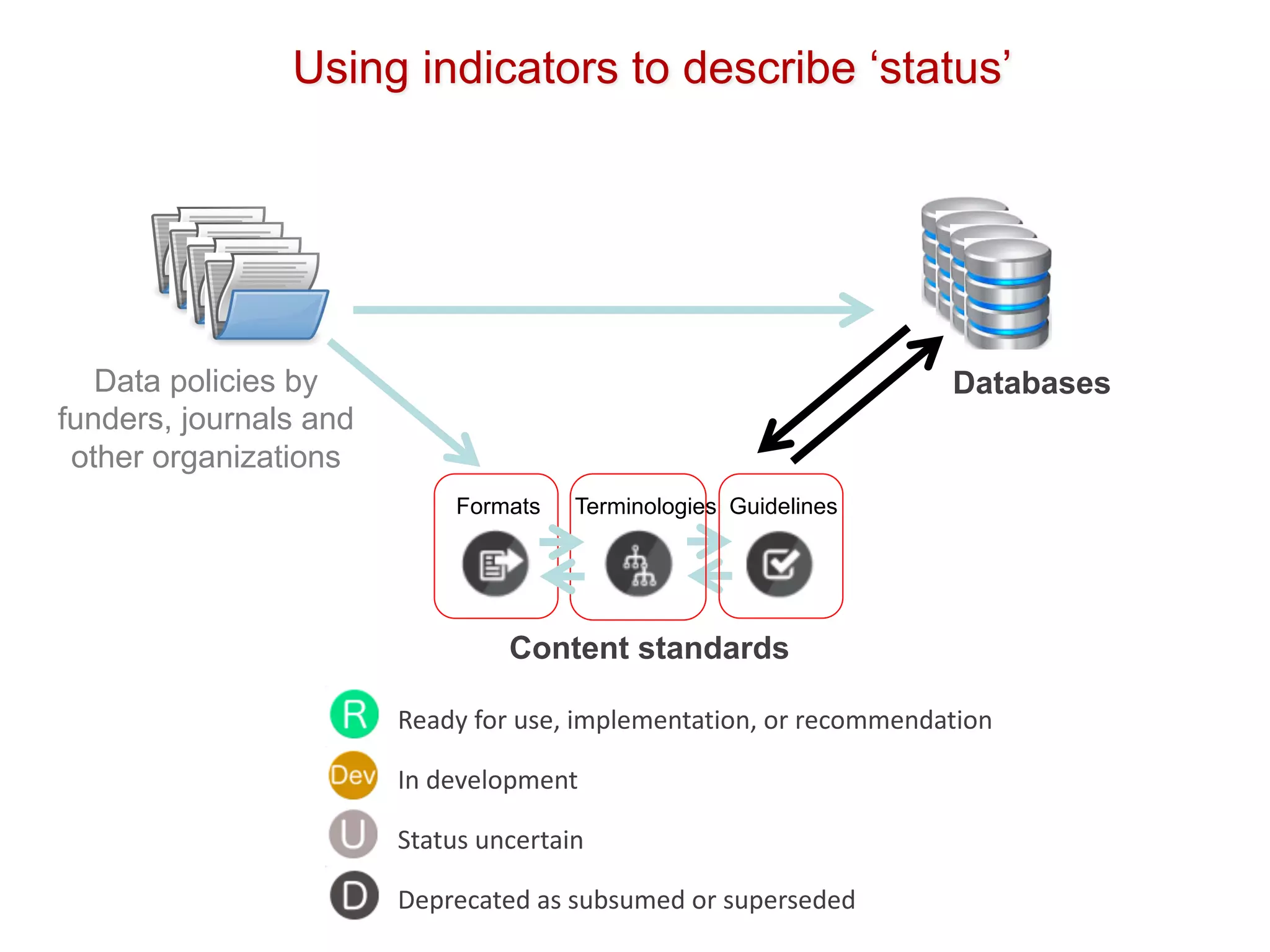
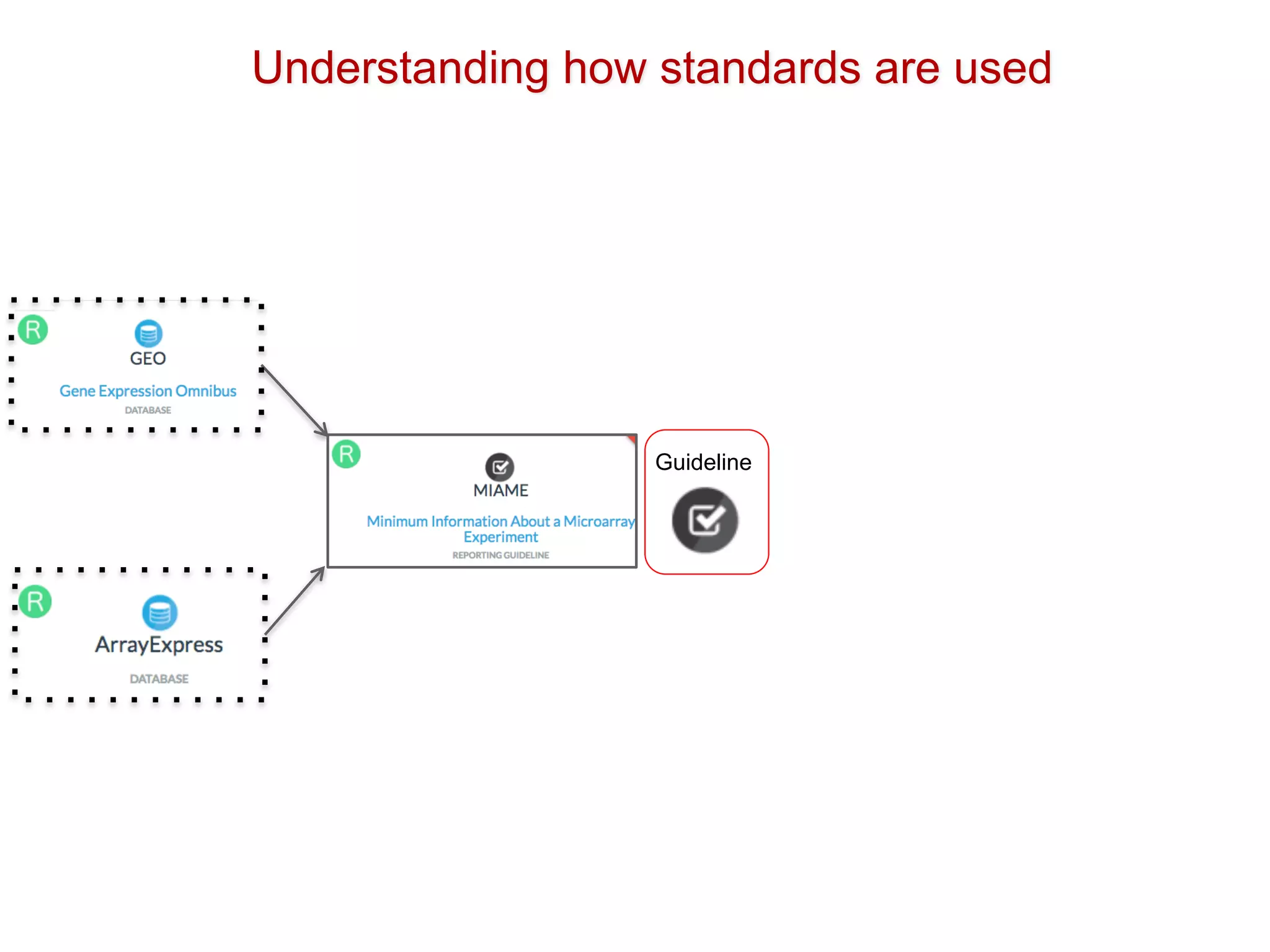
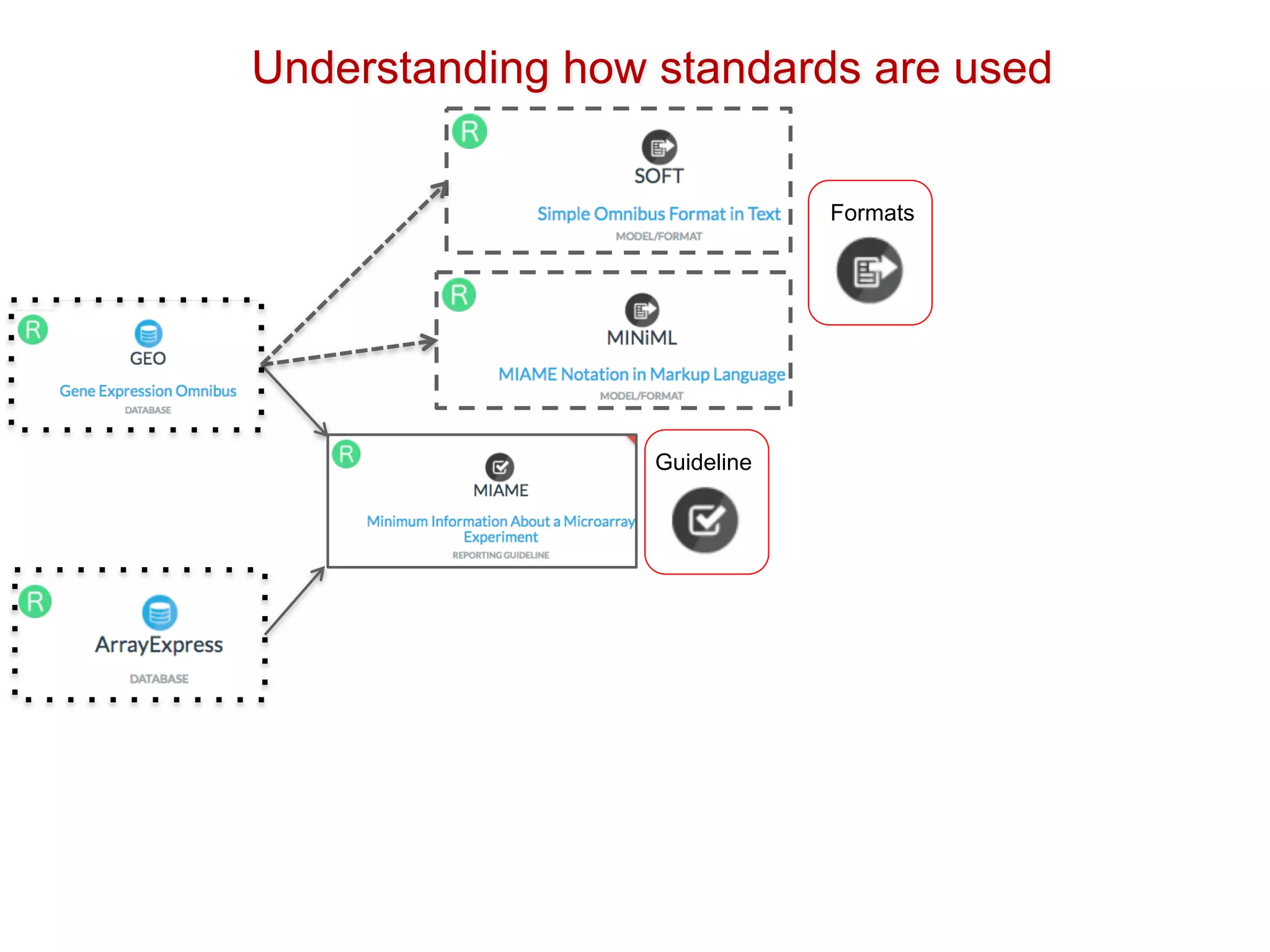
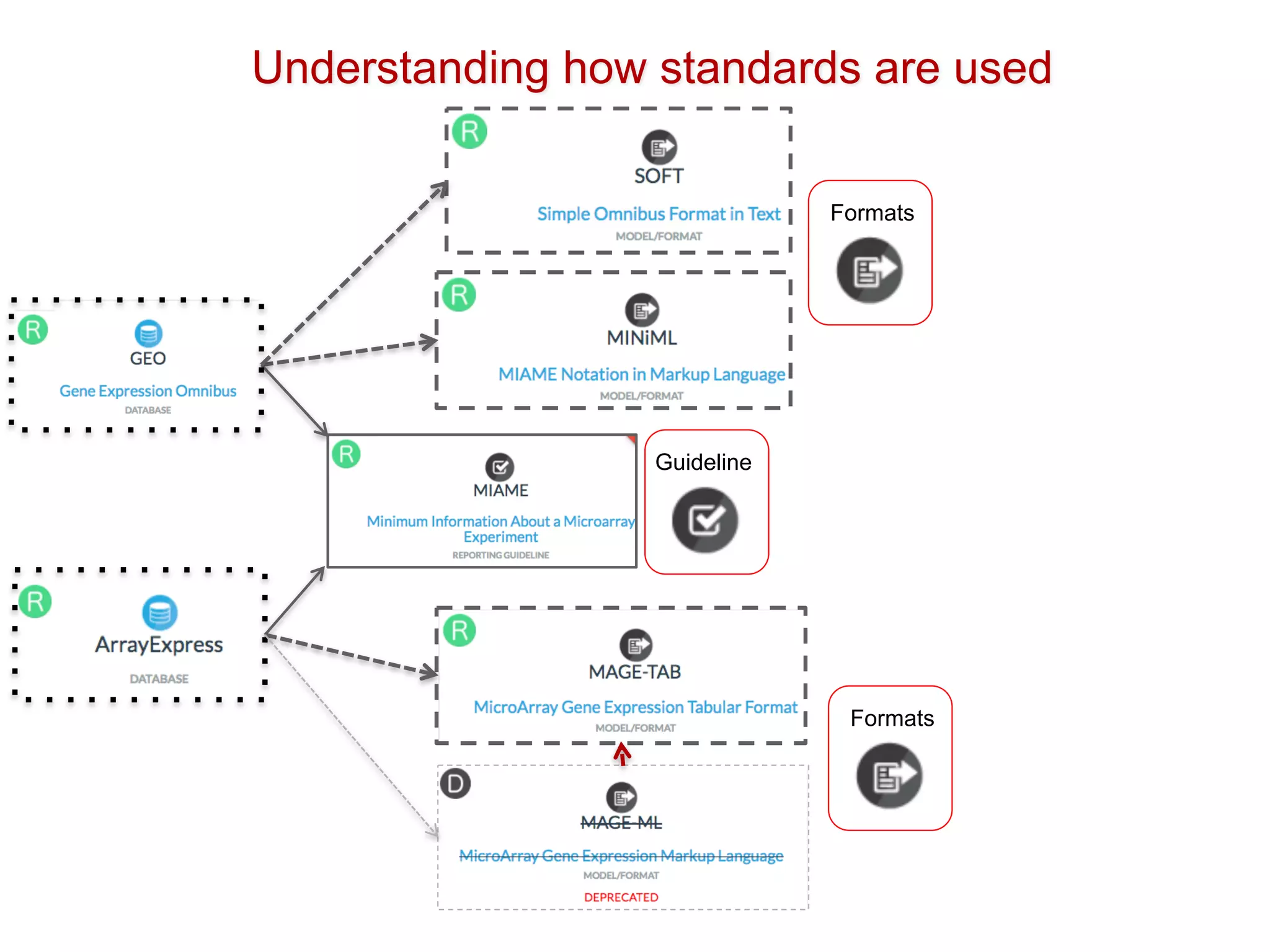
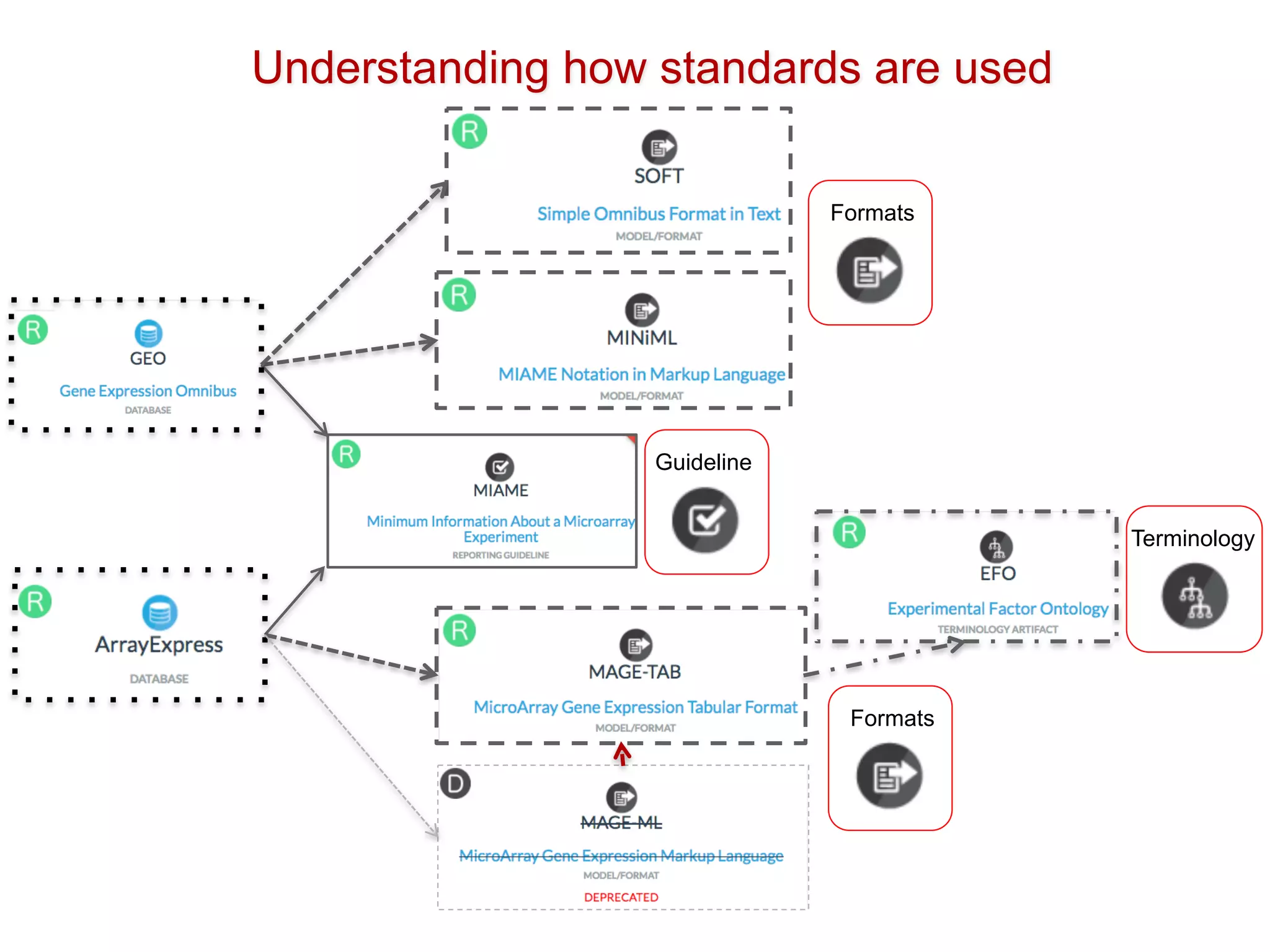
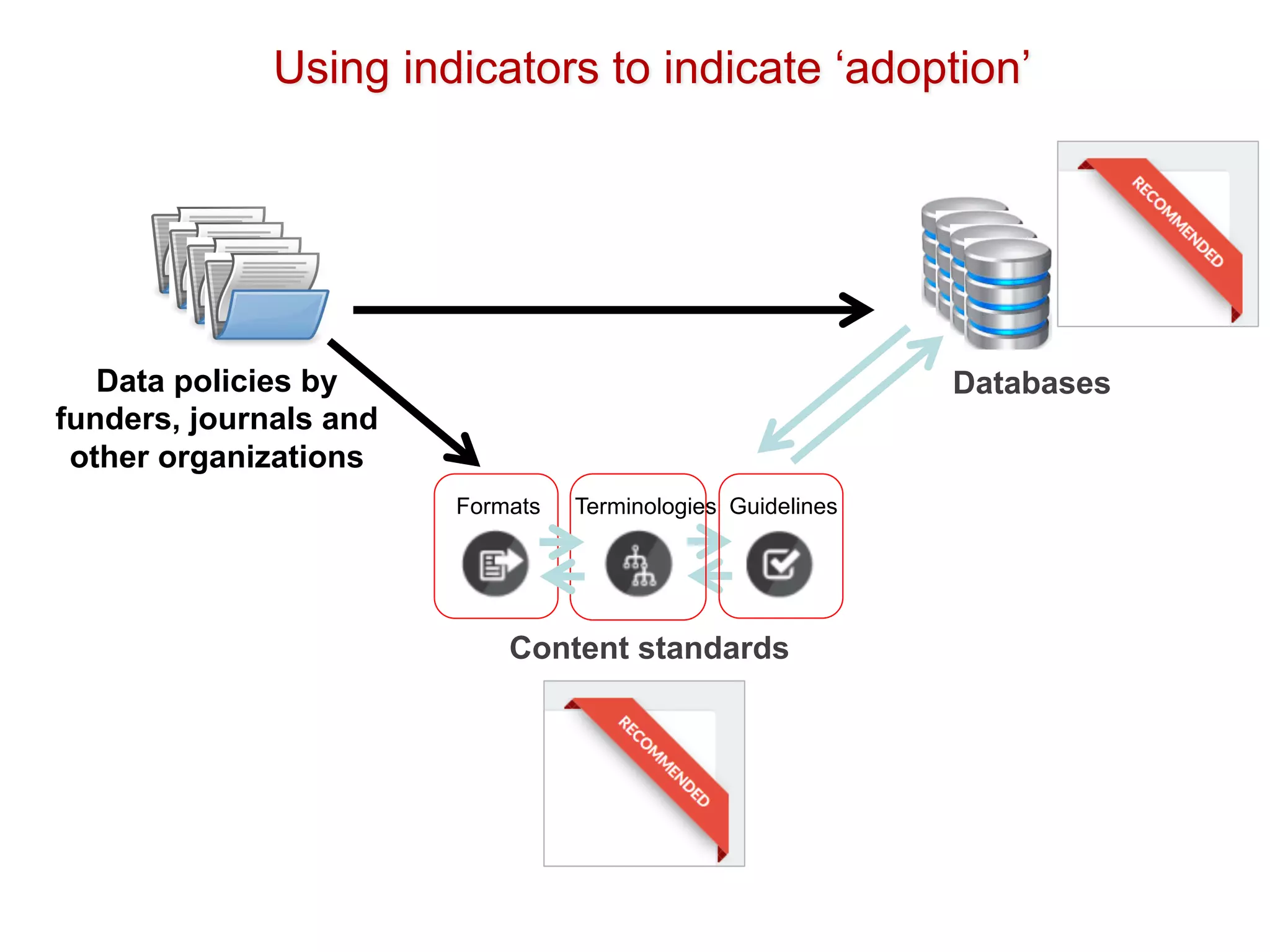
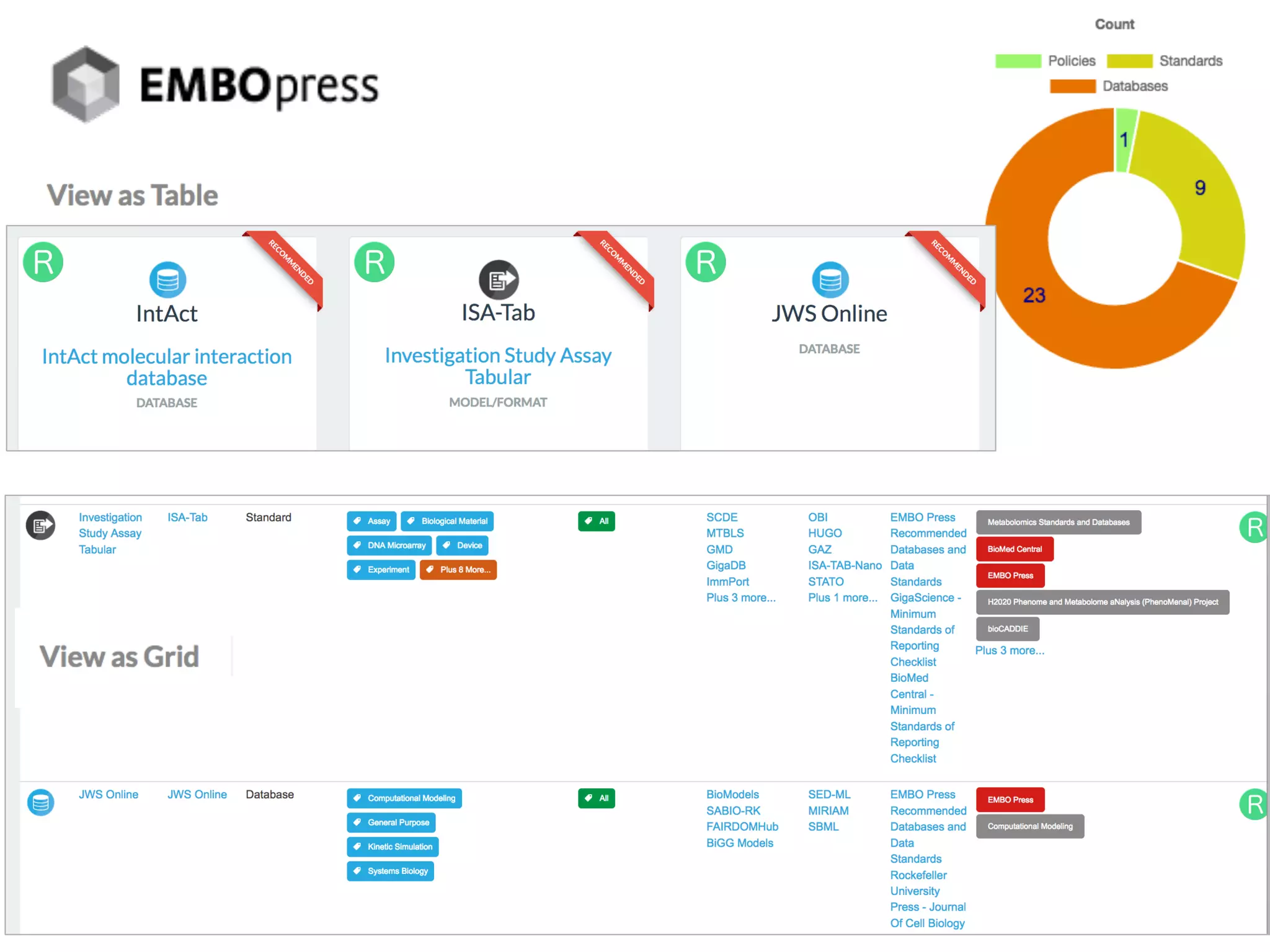
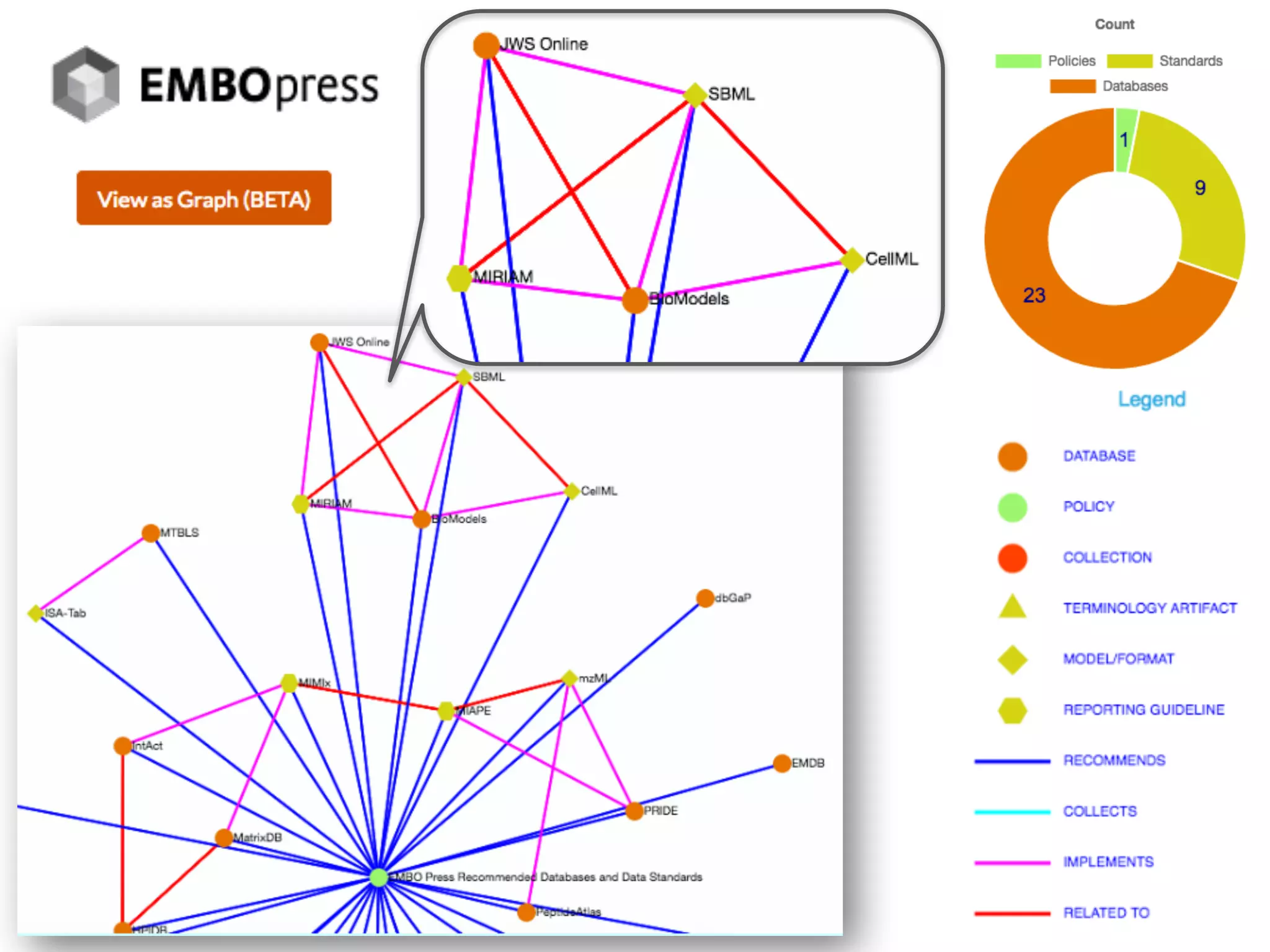
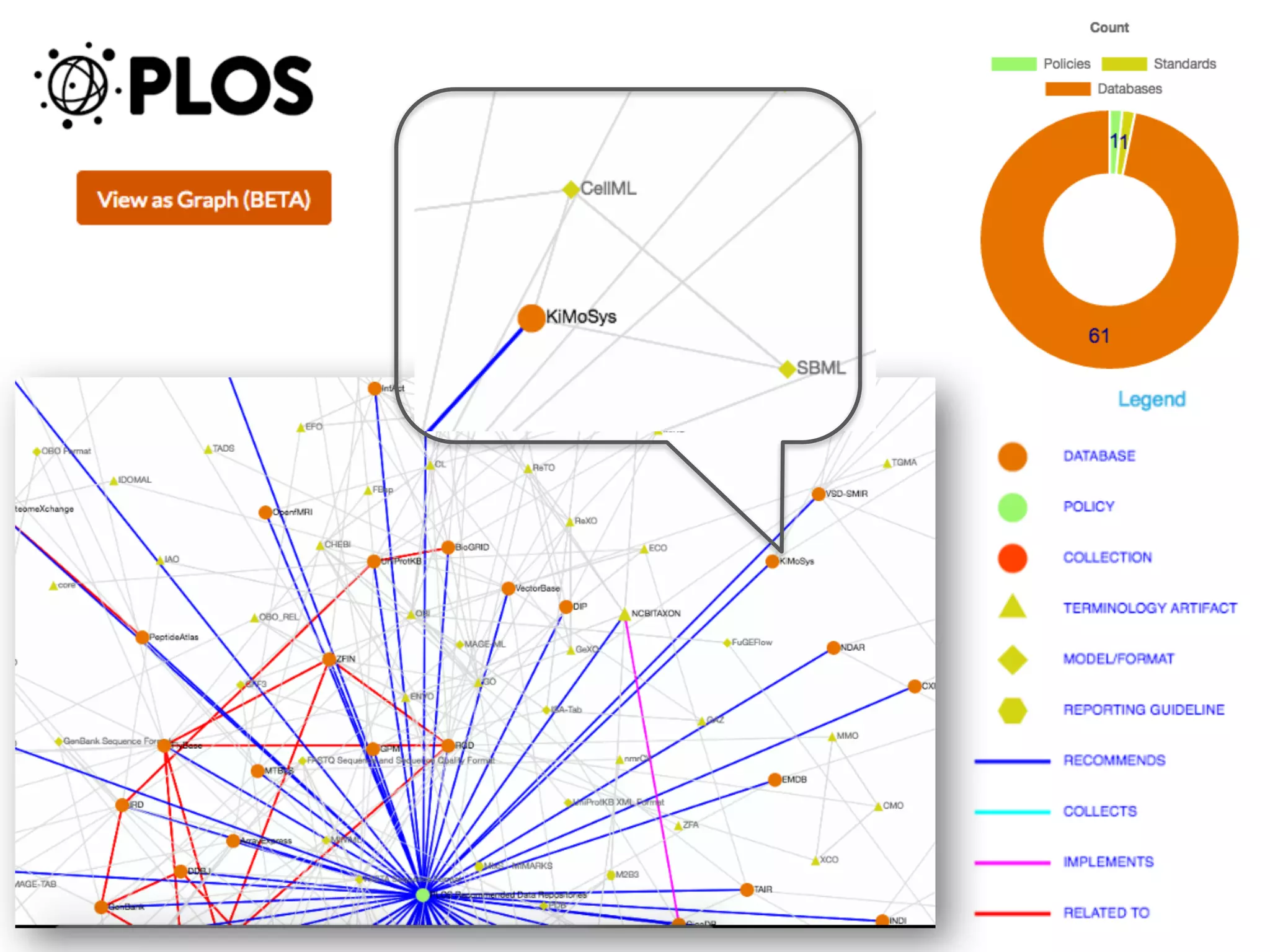
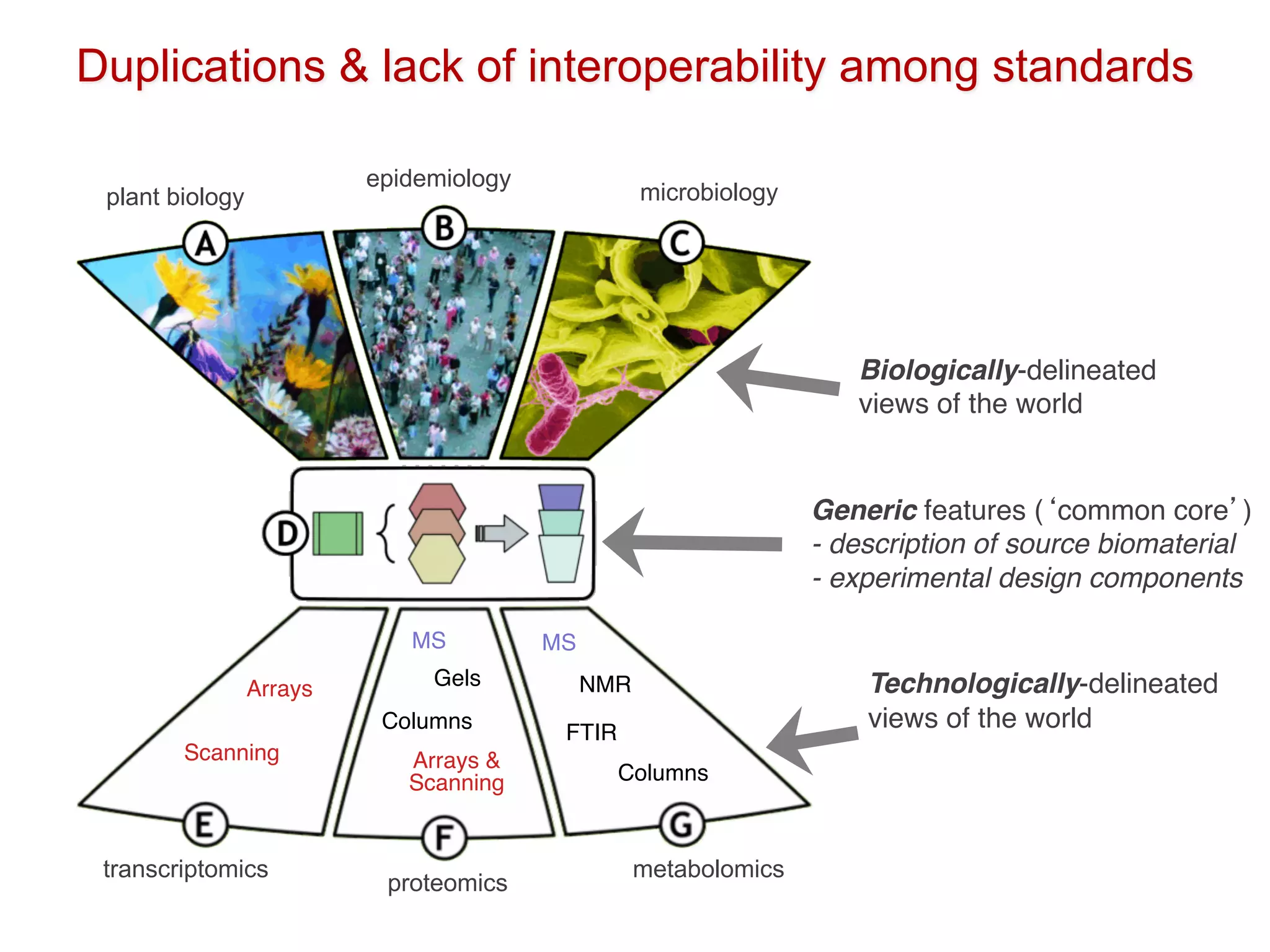
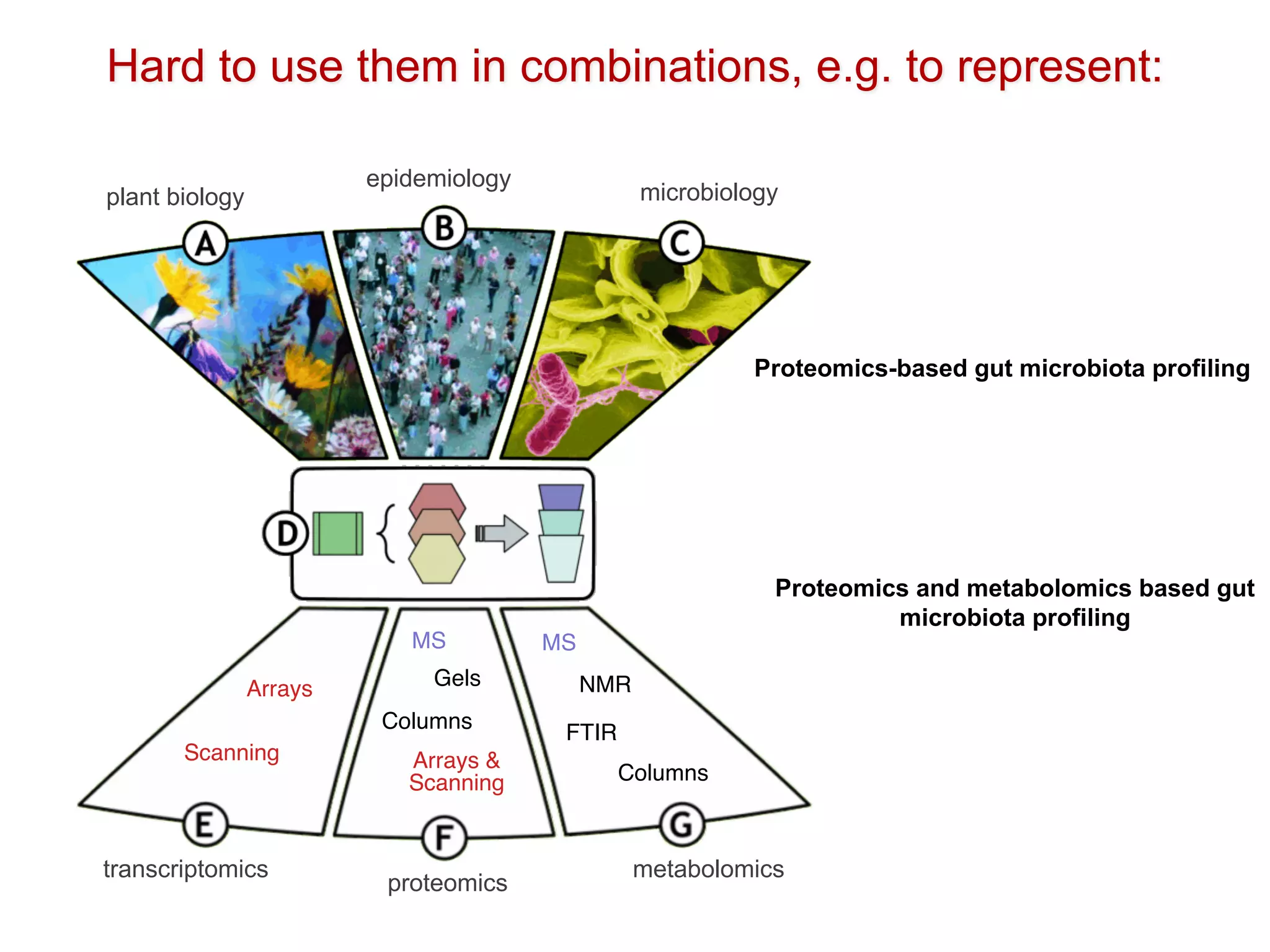
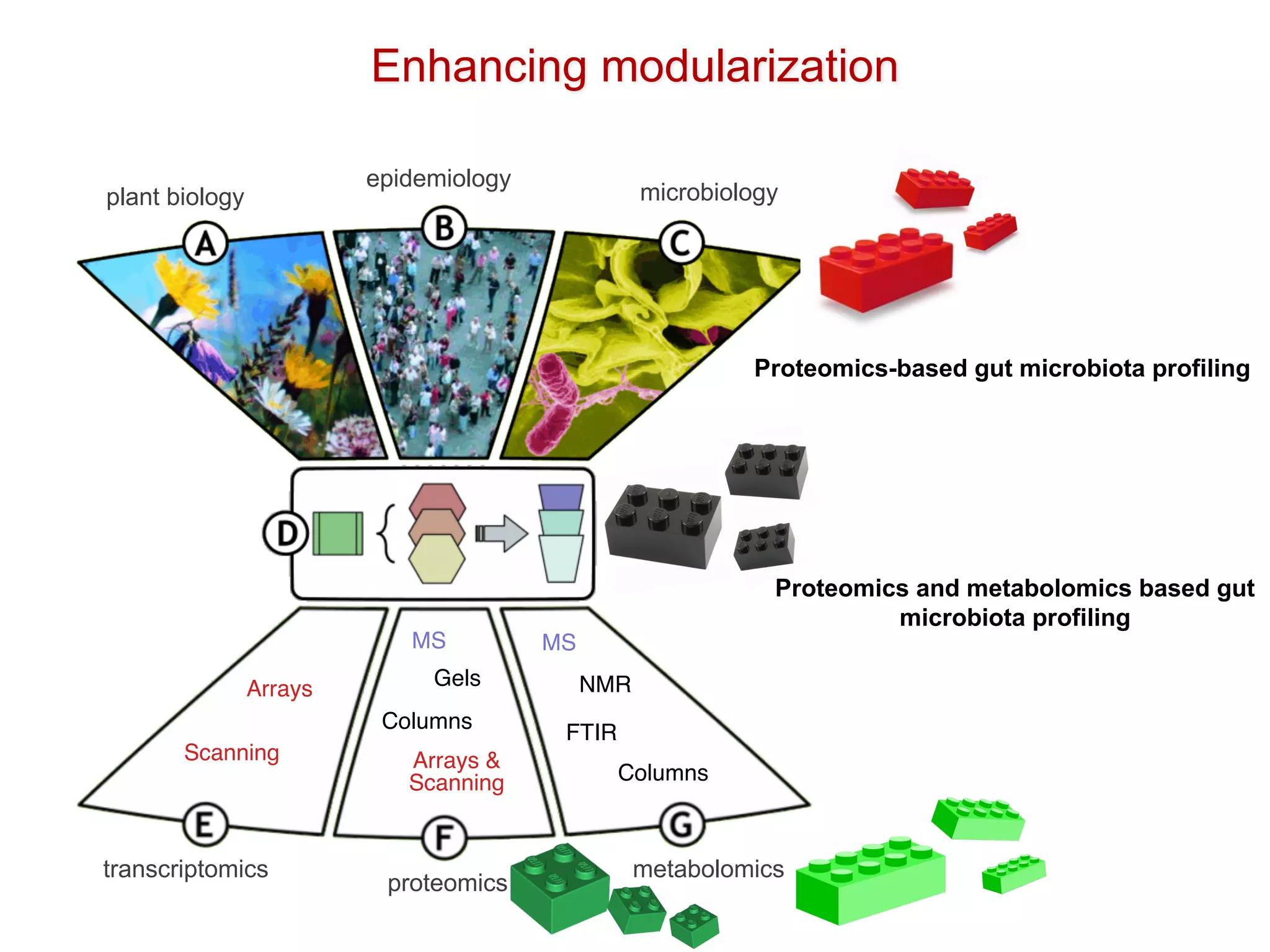
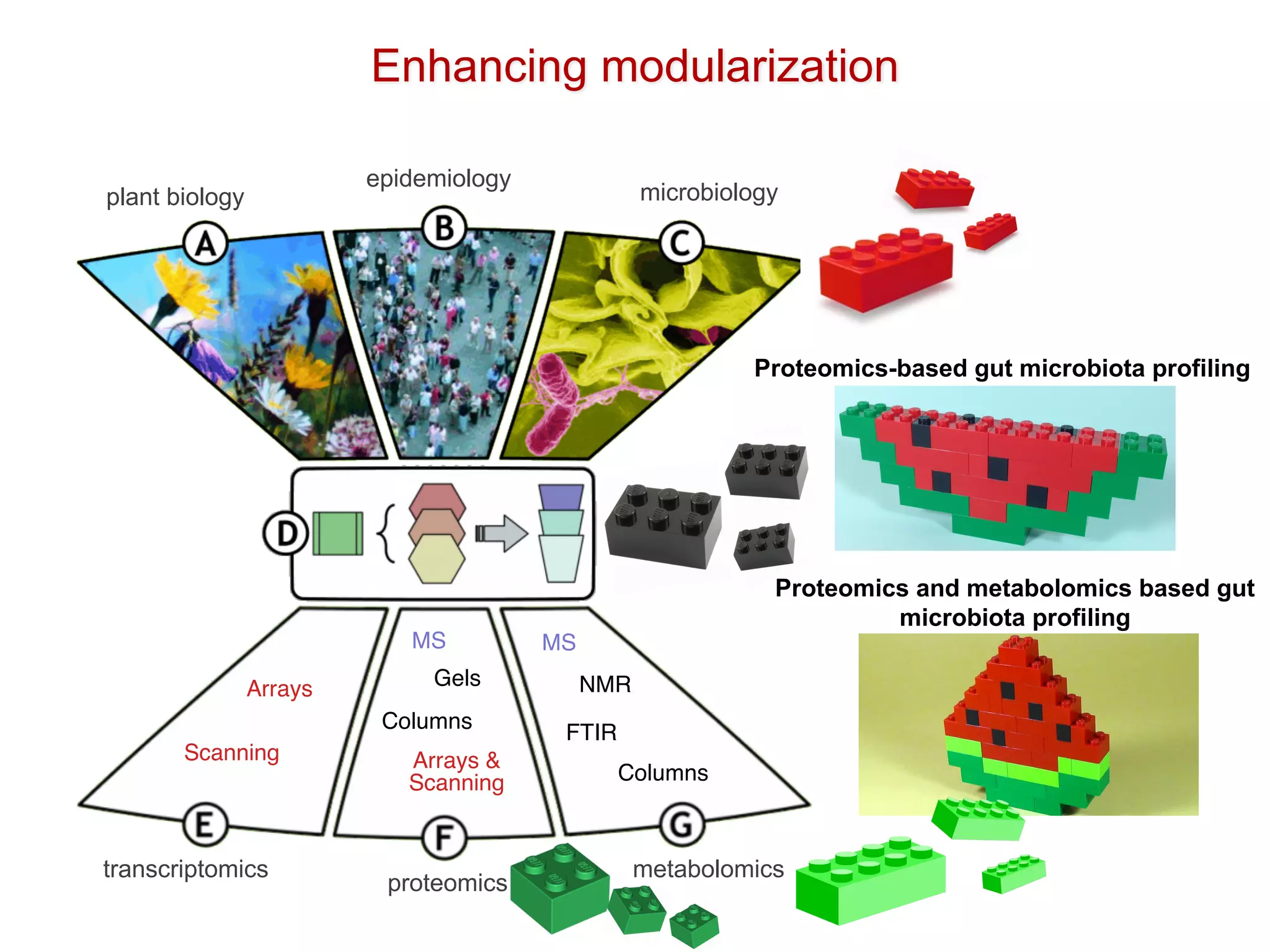
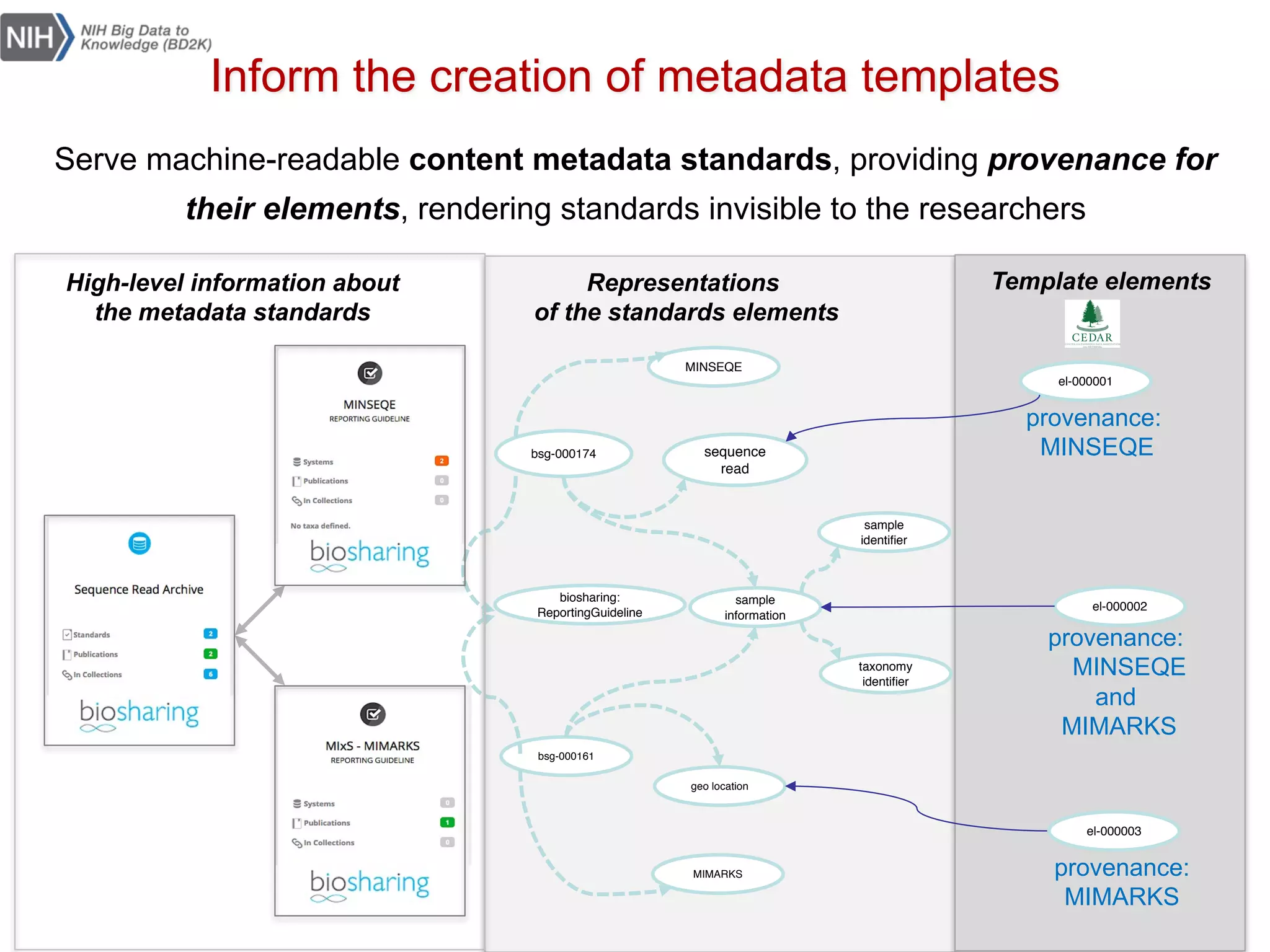
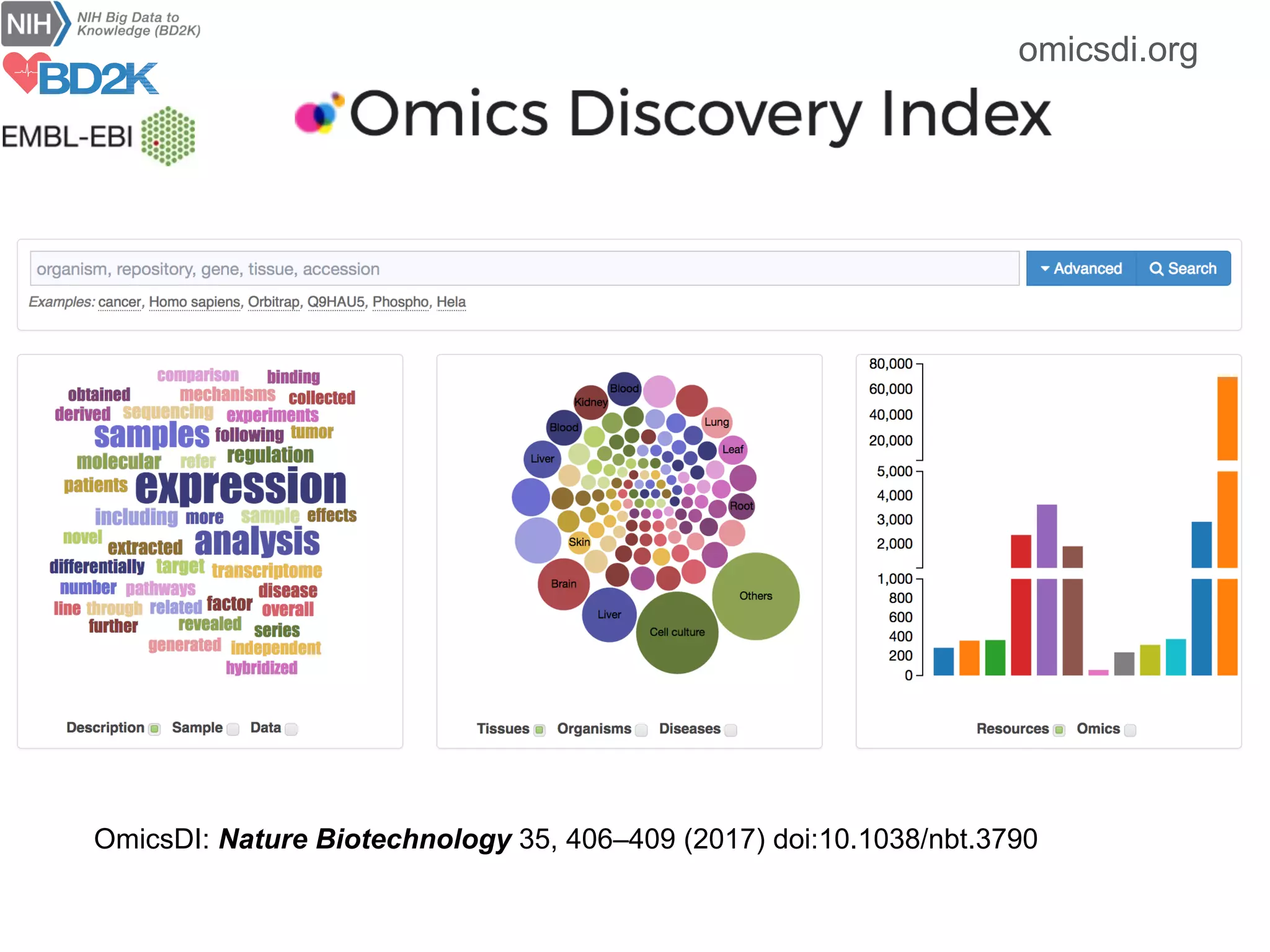
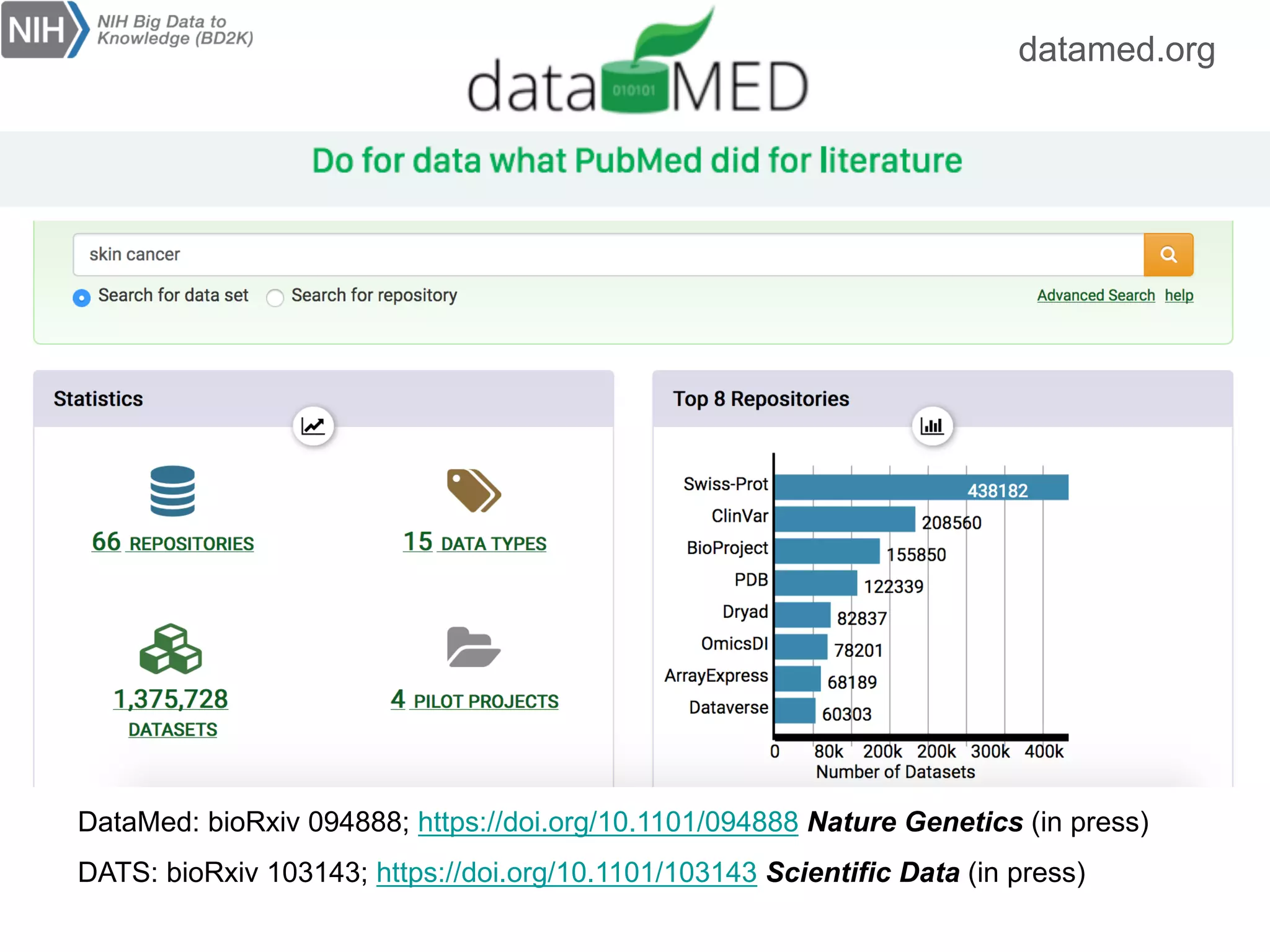
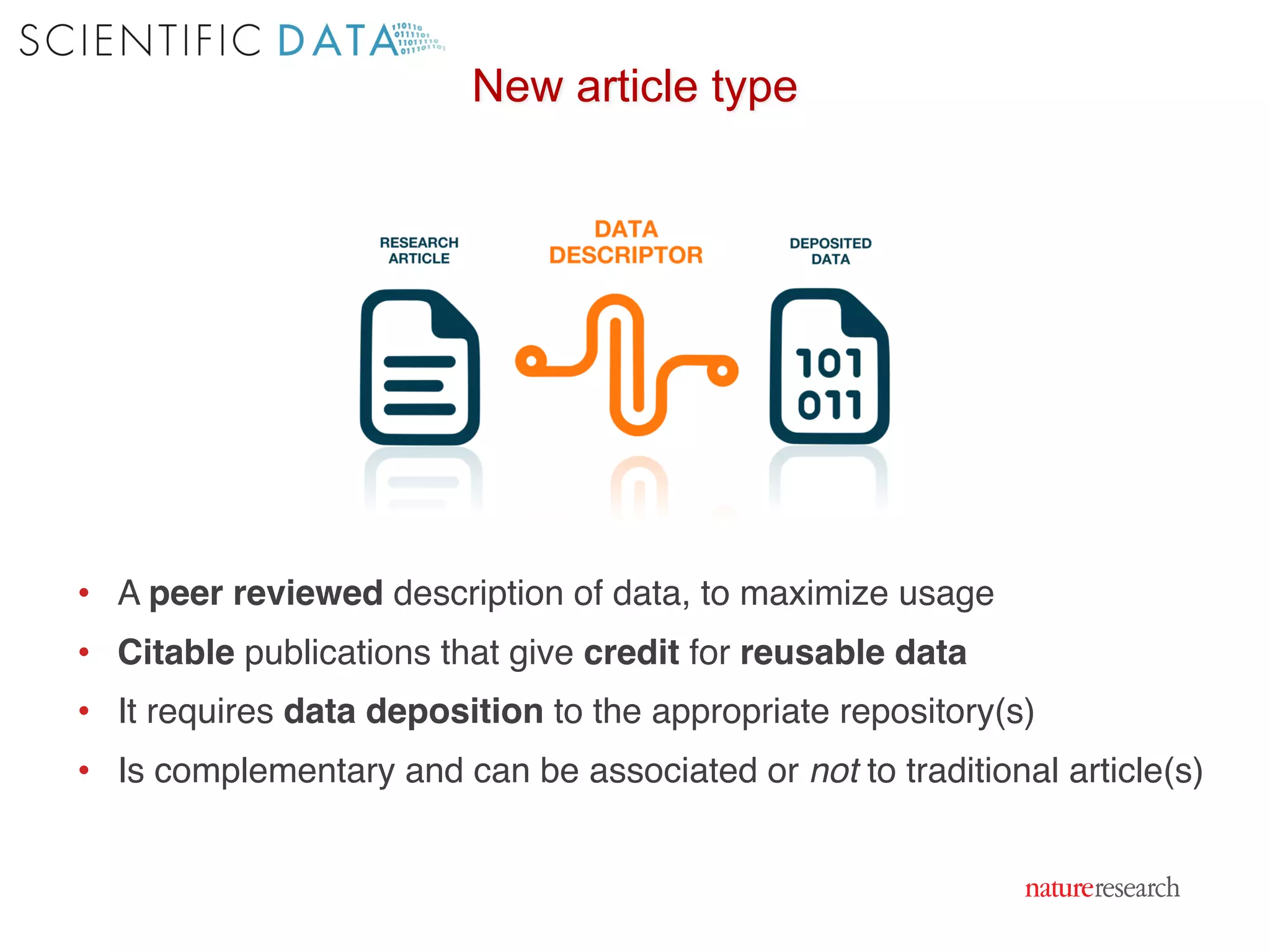
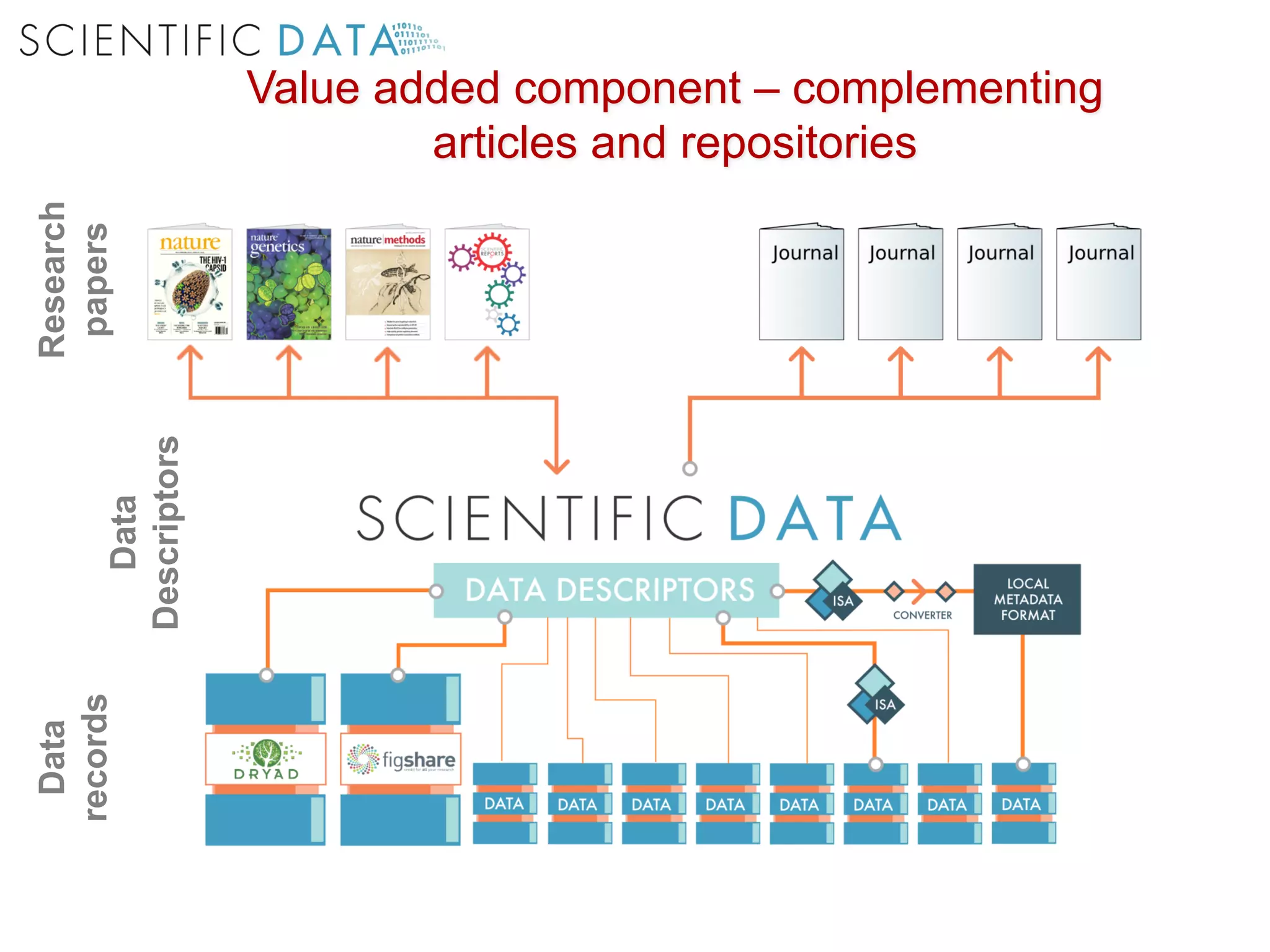
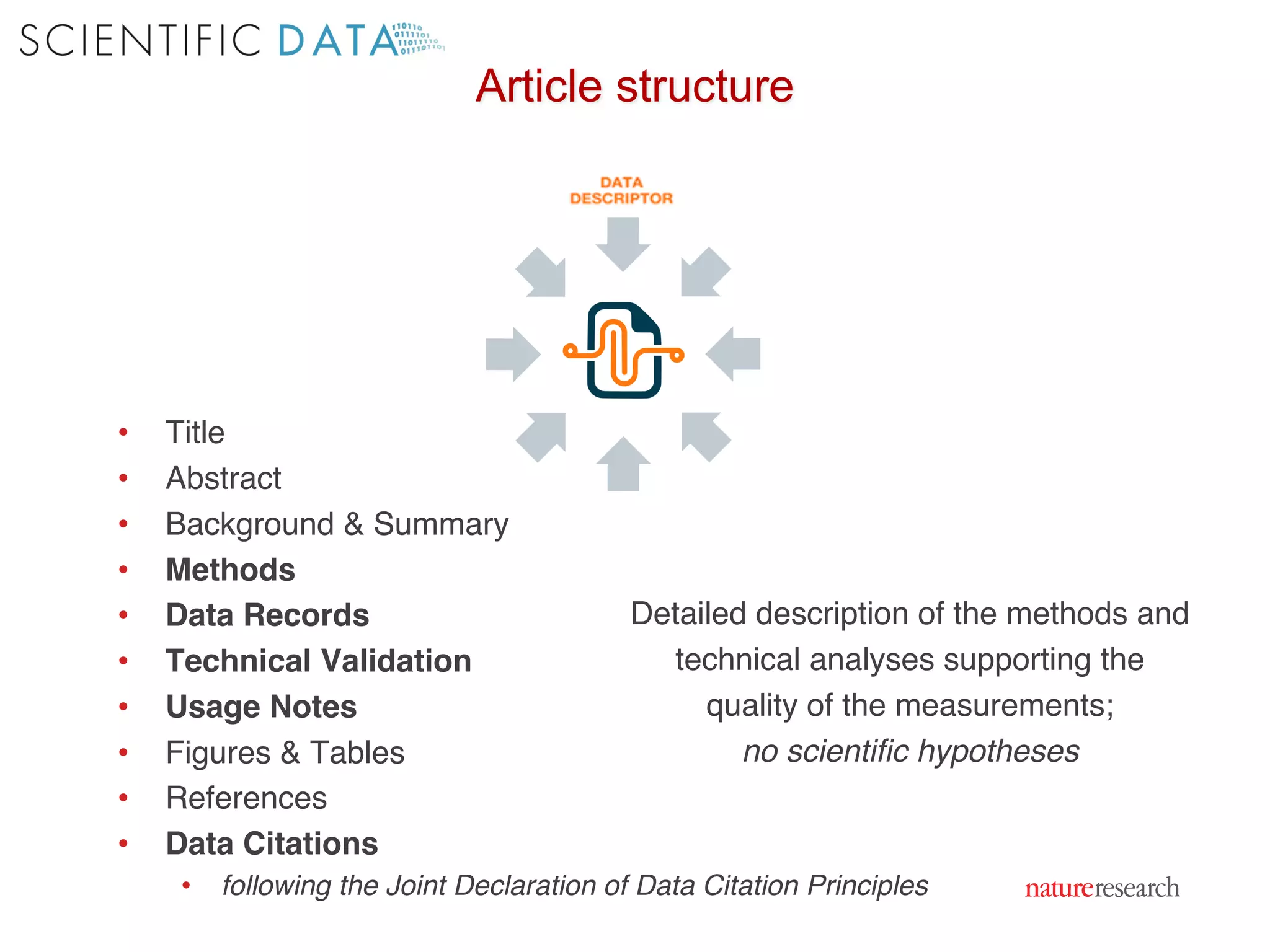
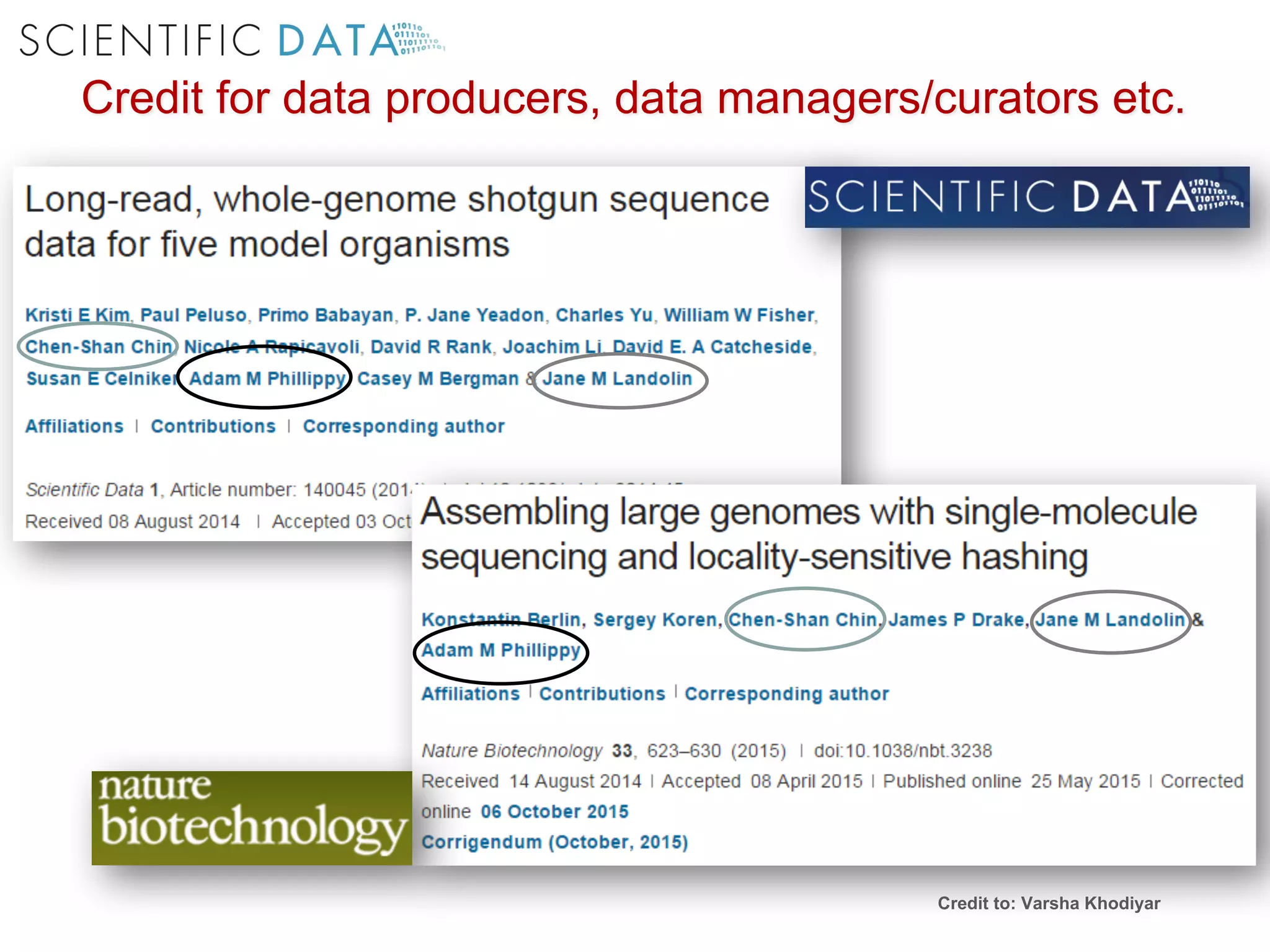
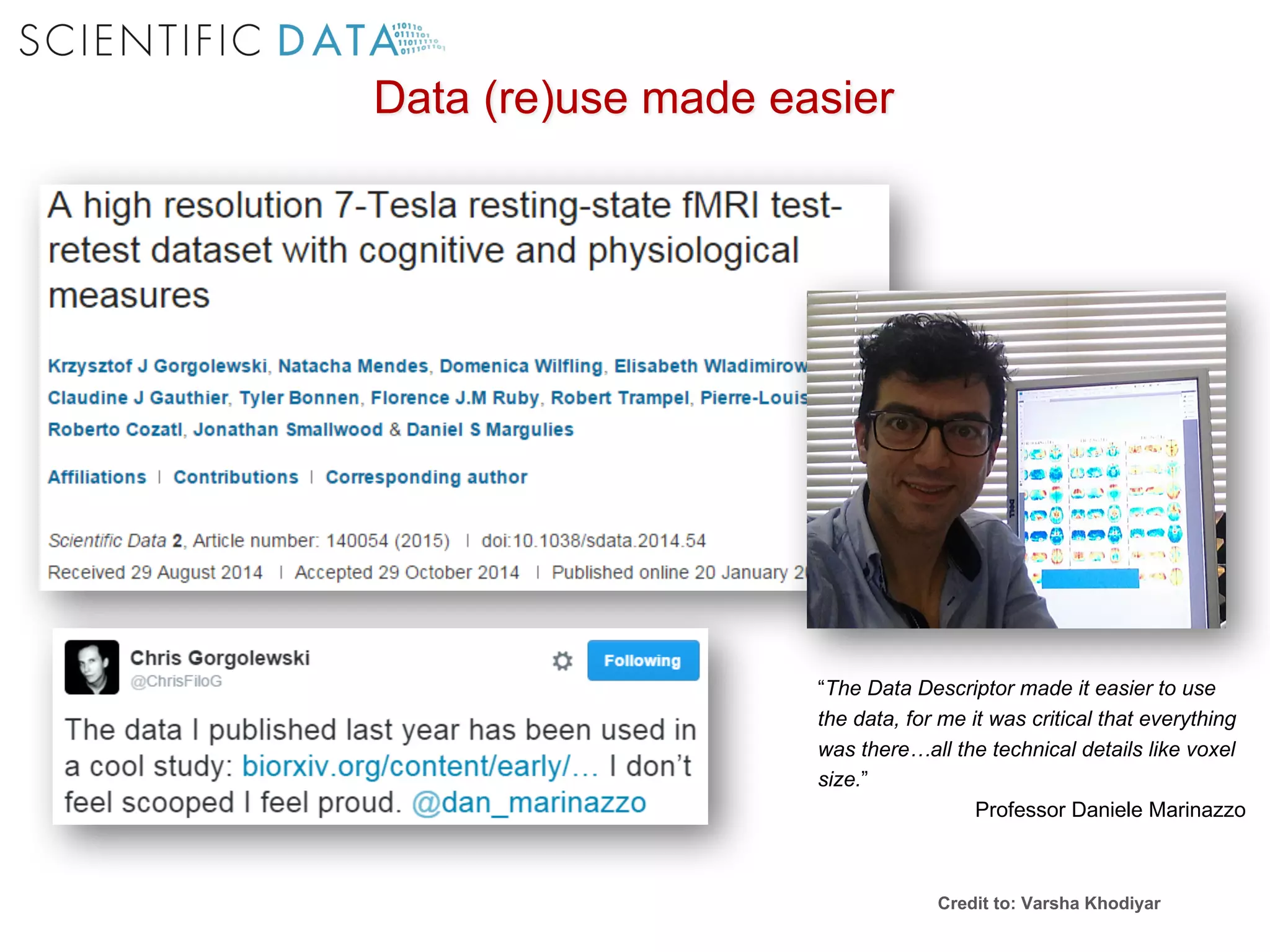
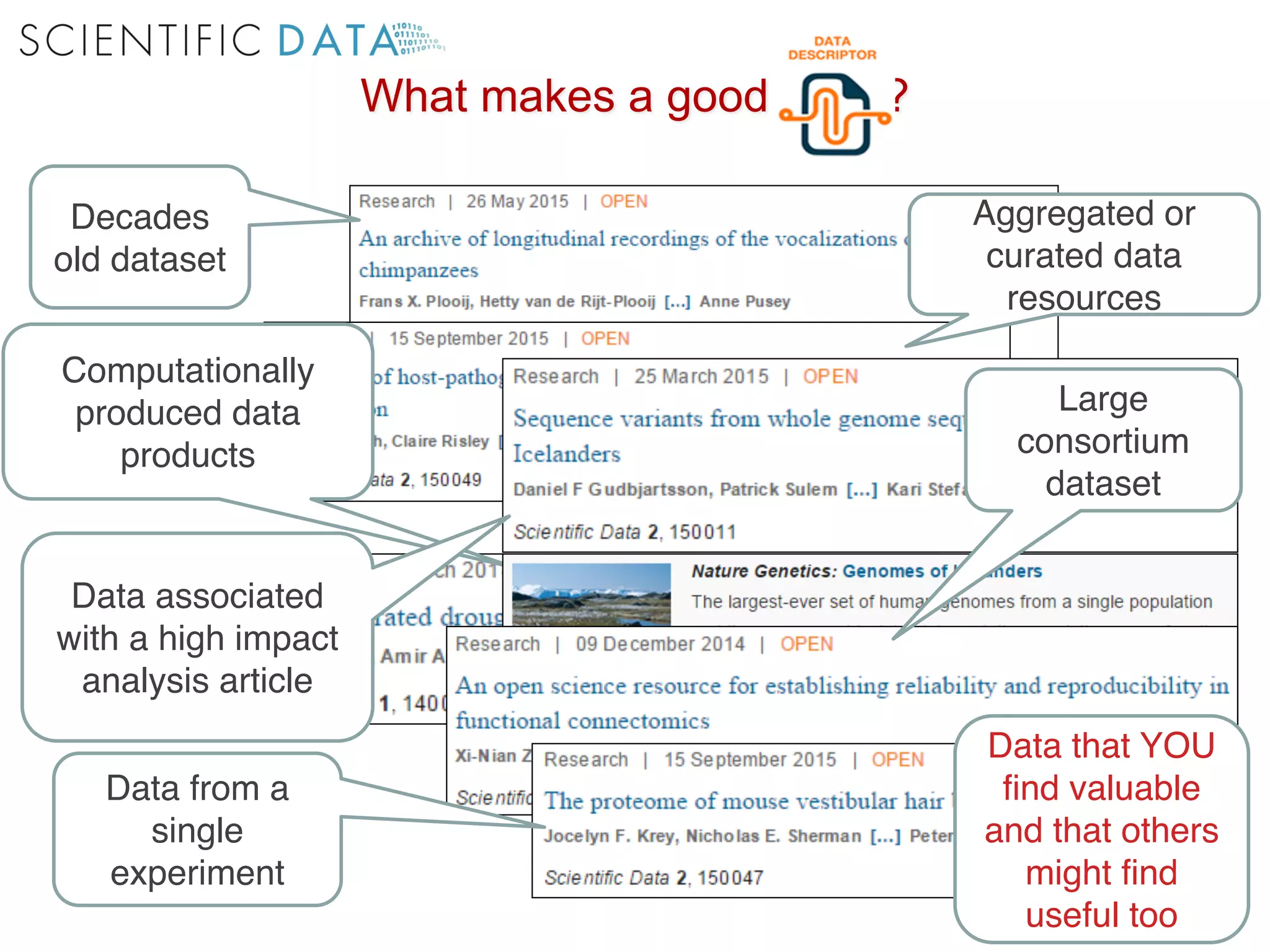
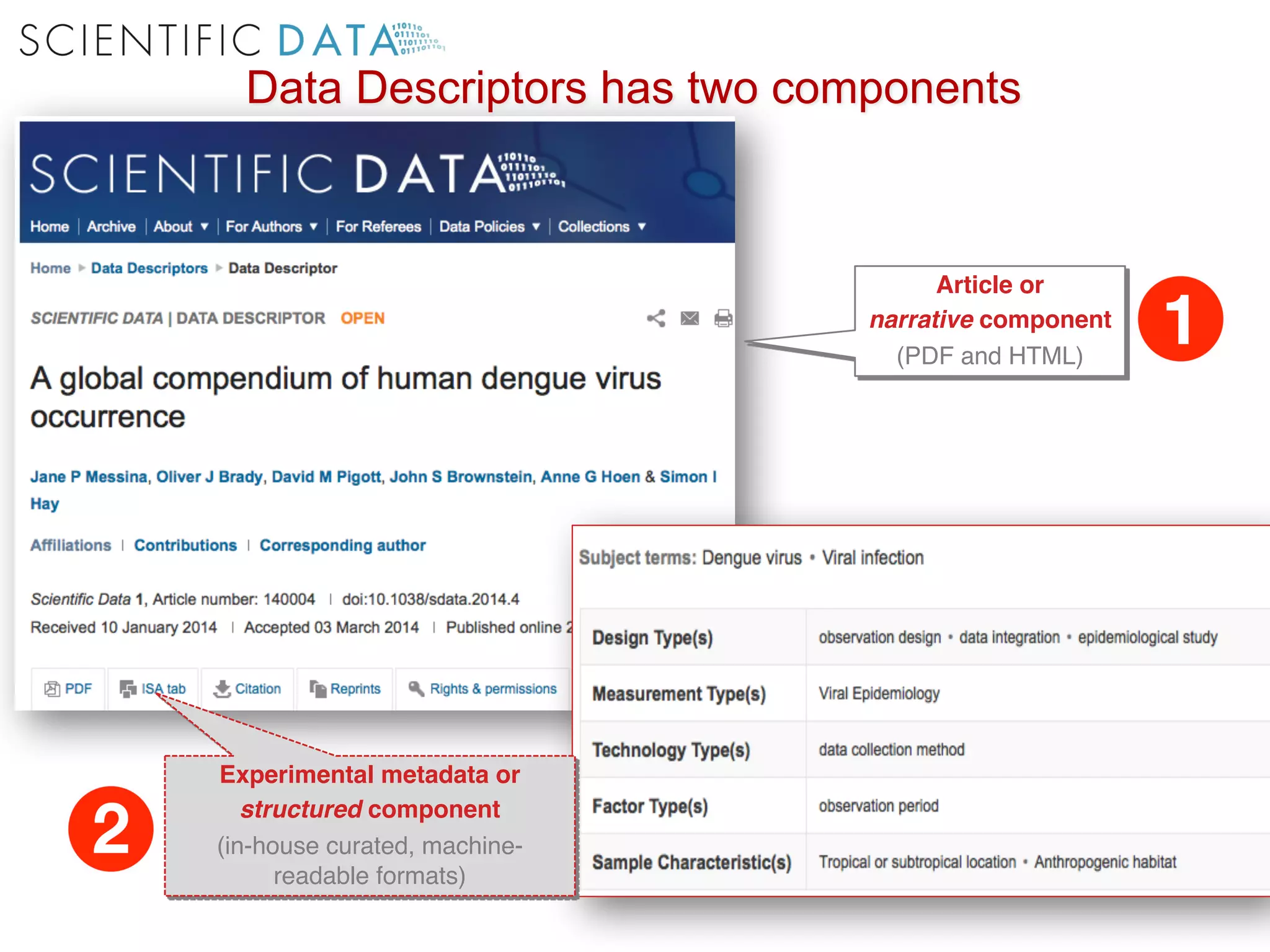
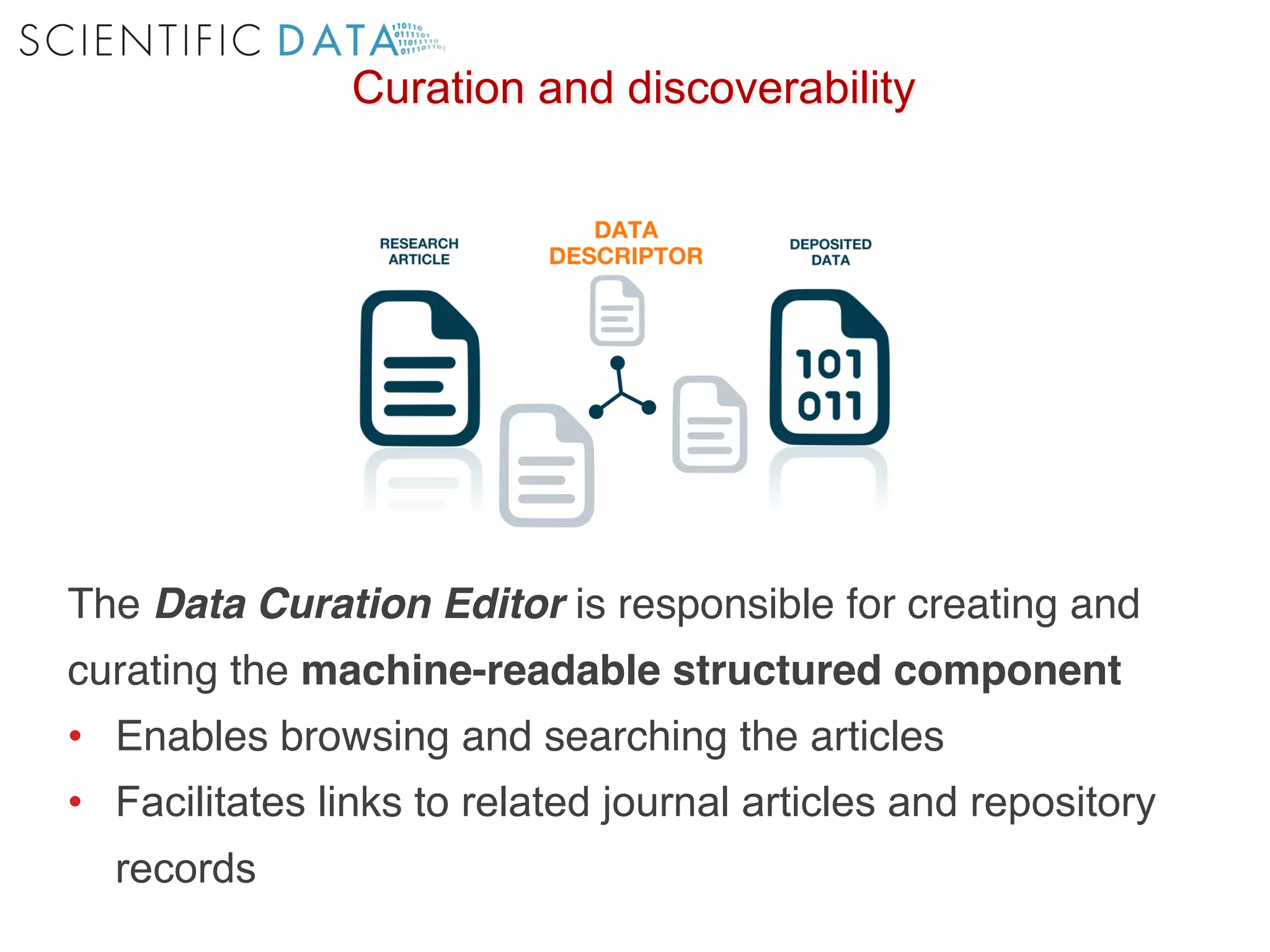
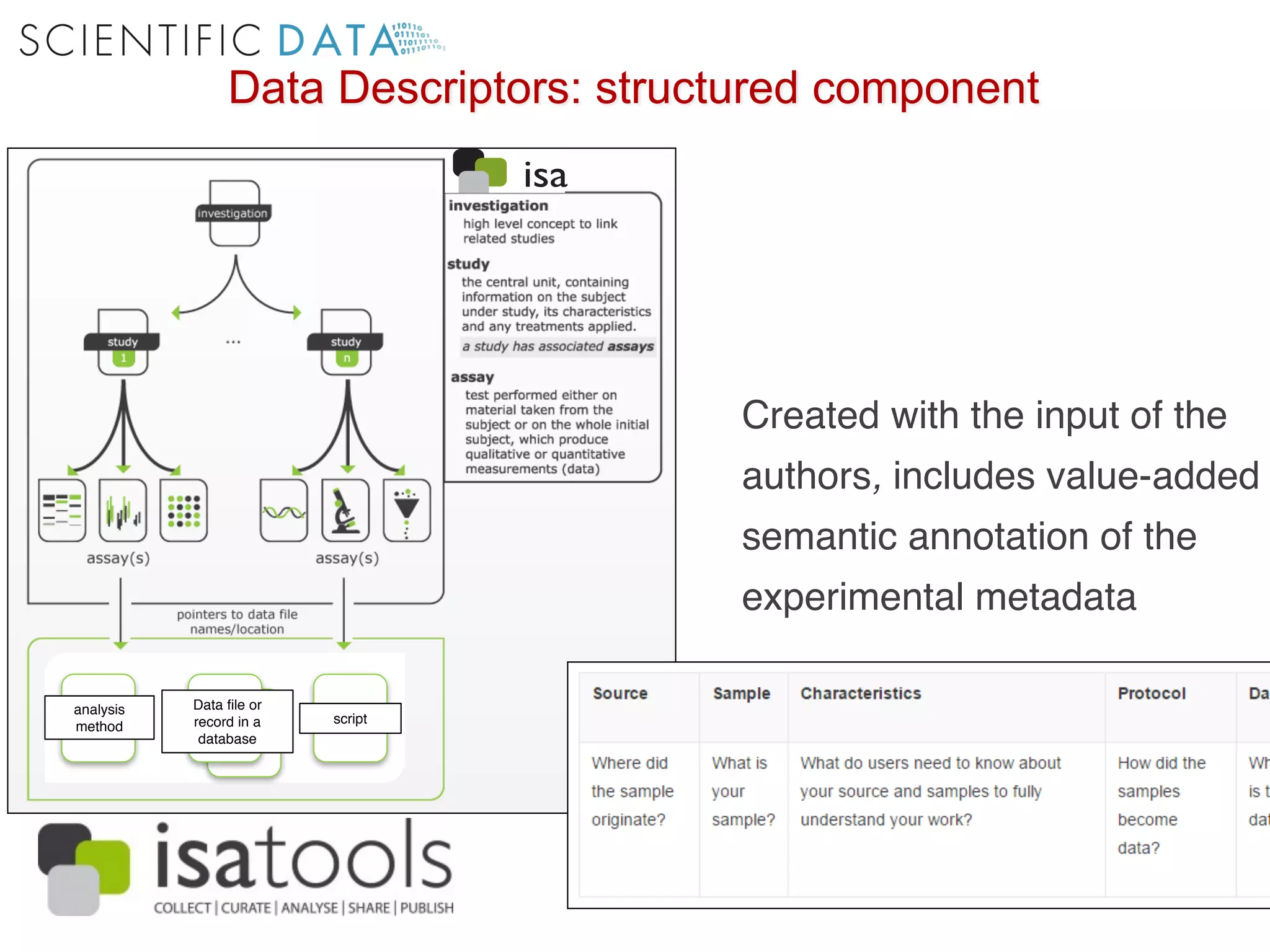
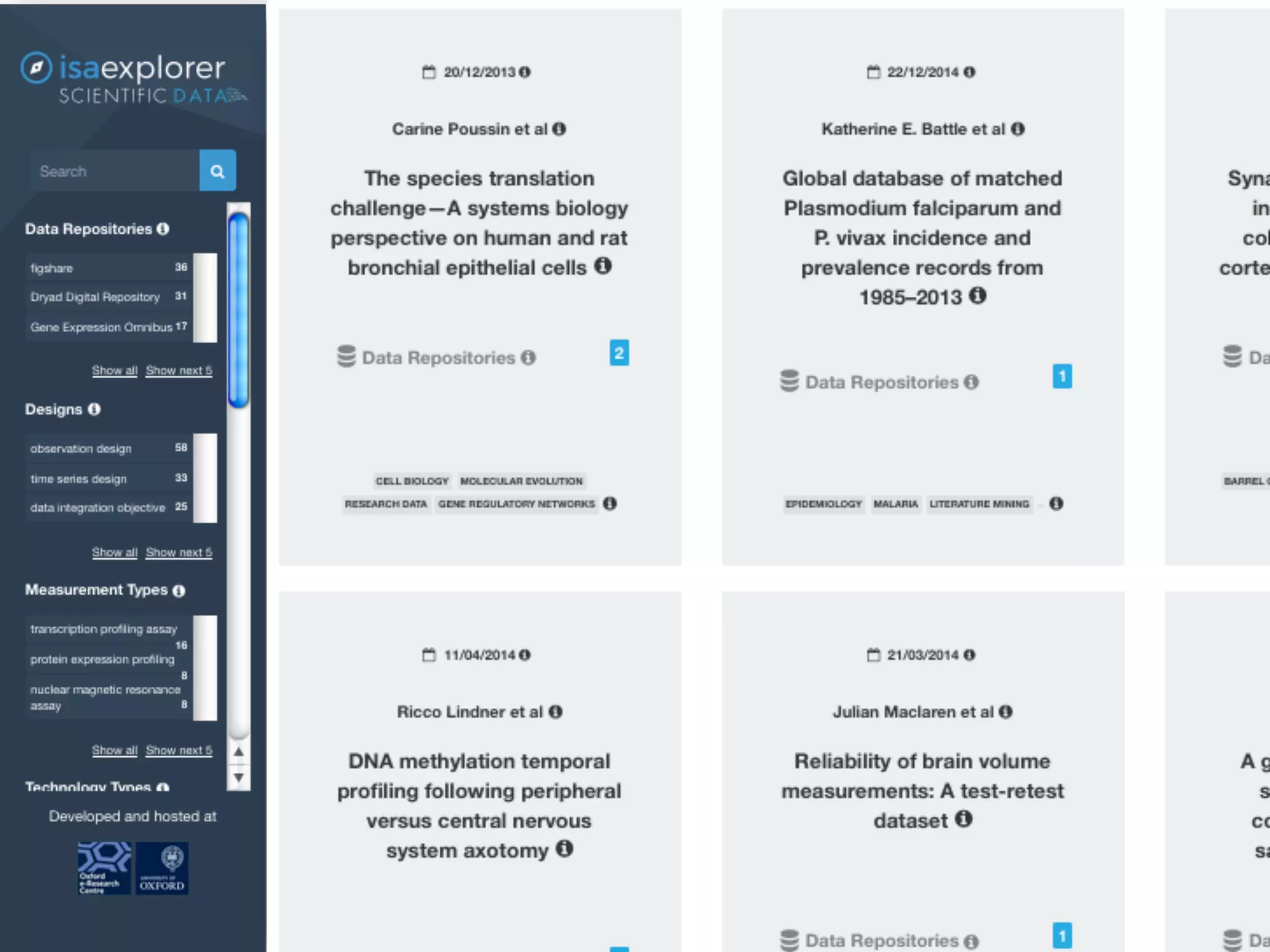
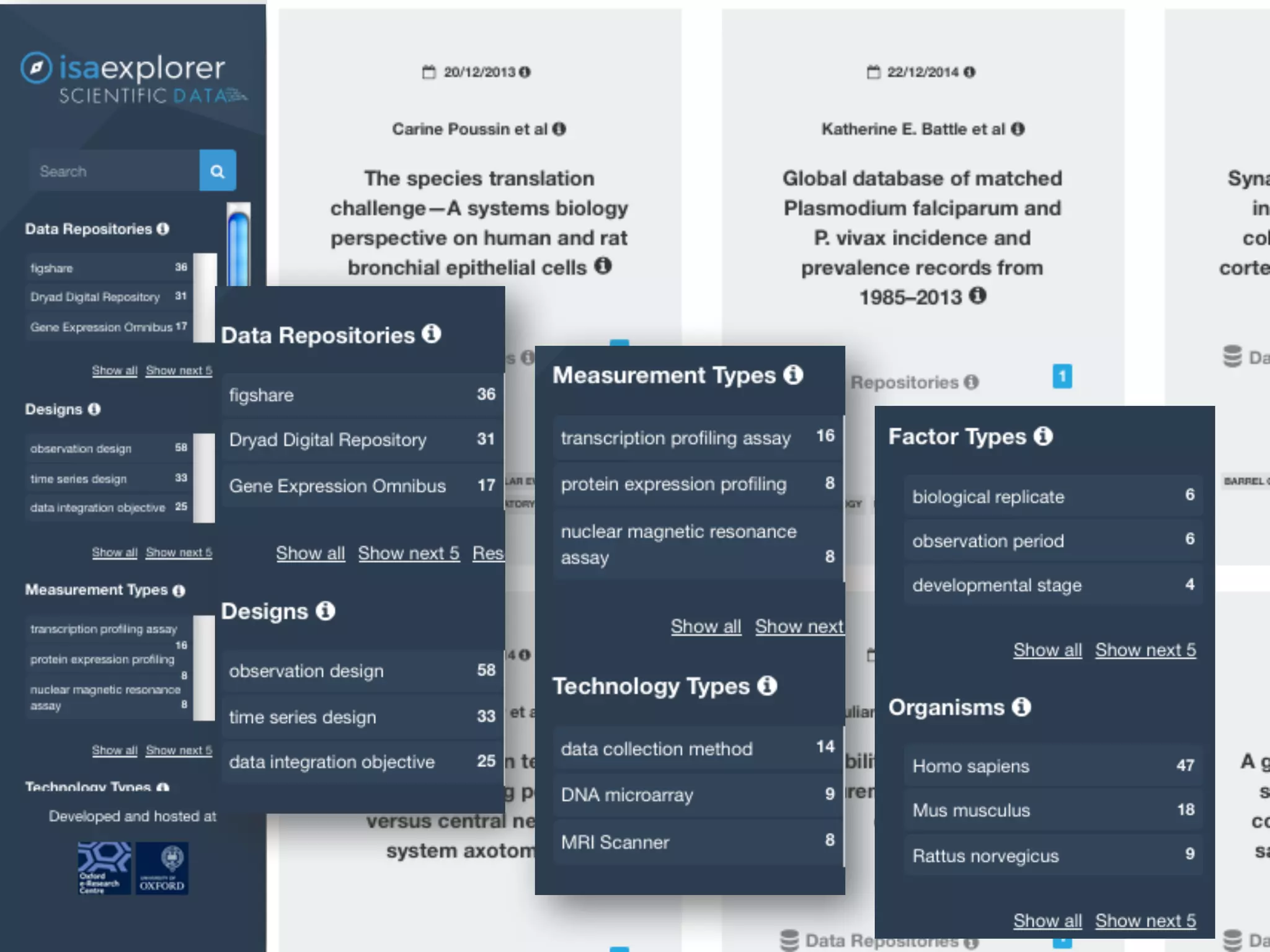
The document discusses the importance of community standards, fair data, and scholarly communication in improving the management and reuse of health data. It highlights key issues such as low findability and understandability of data, the need for content reporting standards, and the role of various guidelines and curated portals in promoting data accessibility and interoperability. The document emphasizes the significance of data descriptors and peer-reviewed data publications in enhancing the discoverability and reusability of scientific datasets.