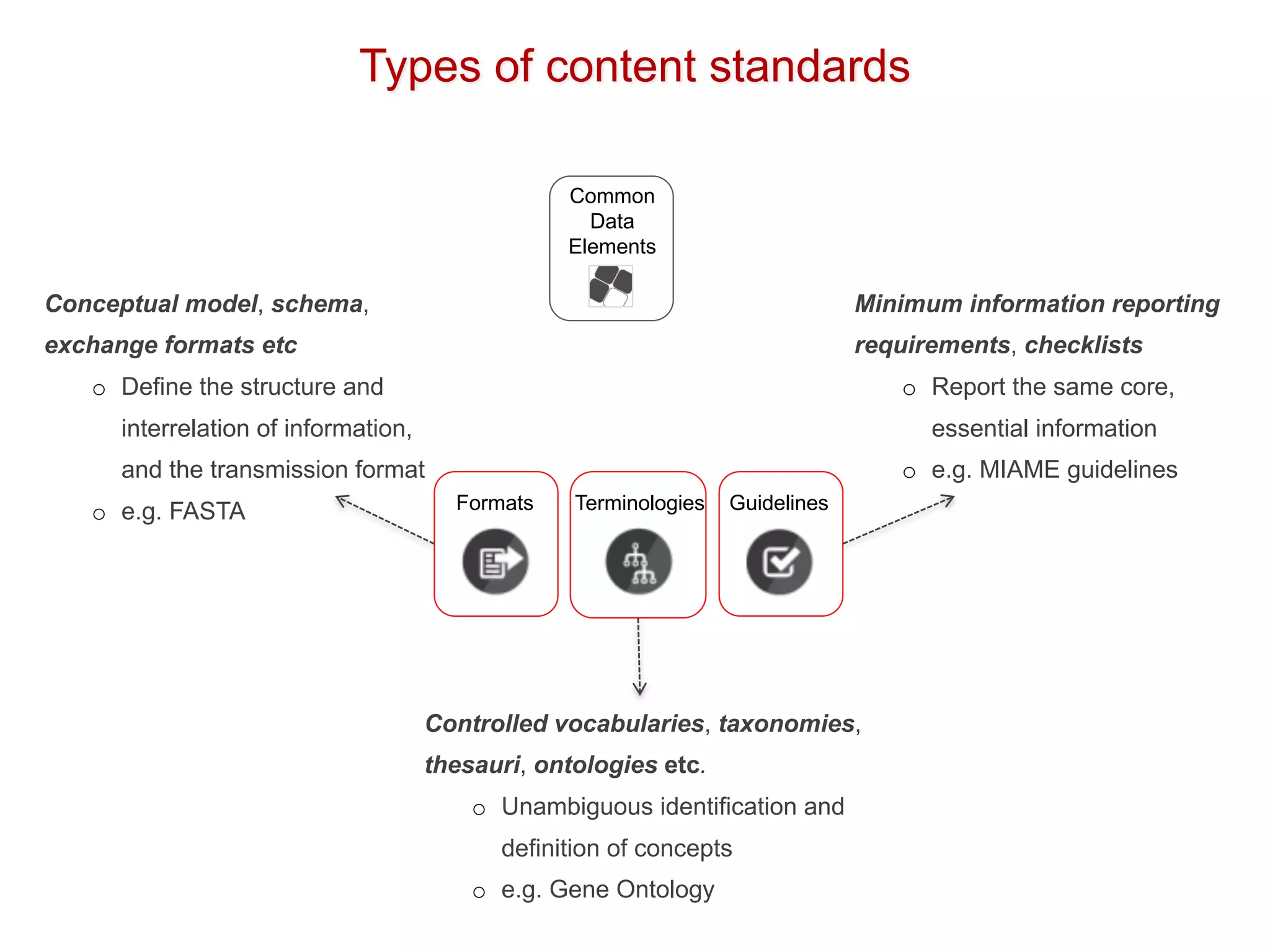
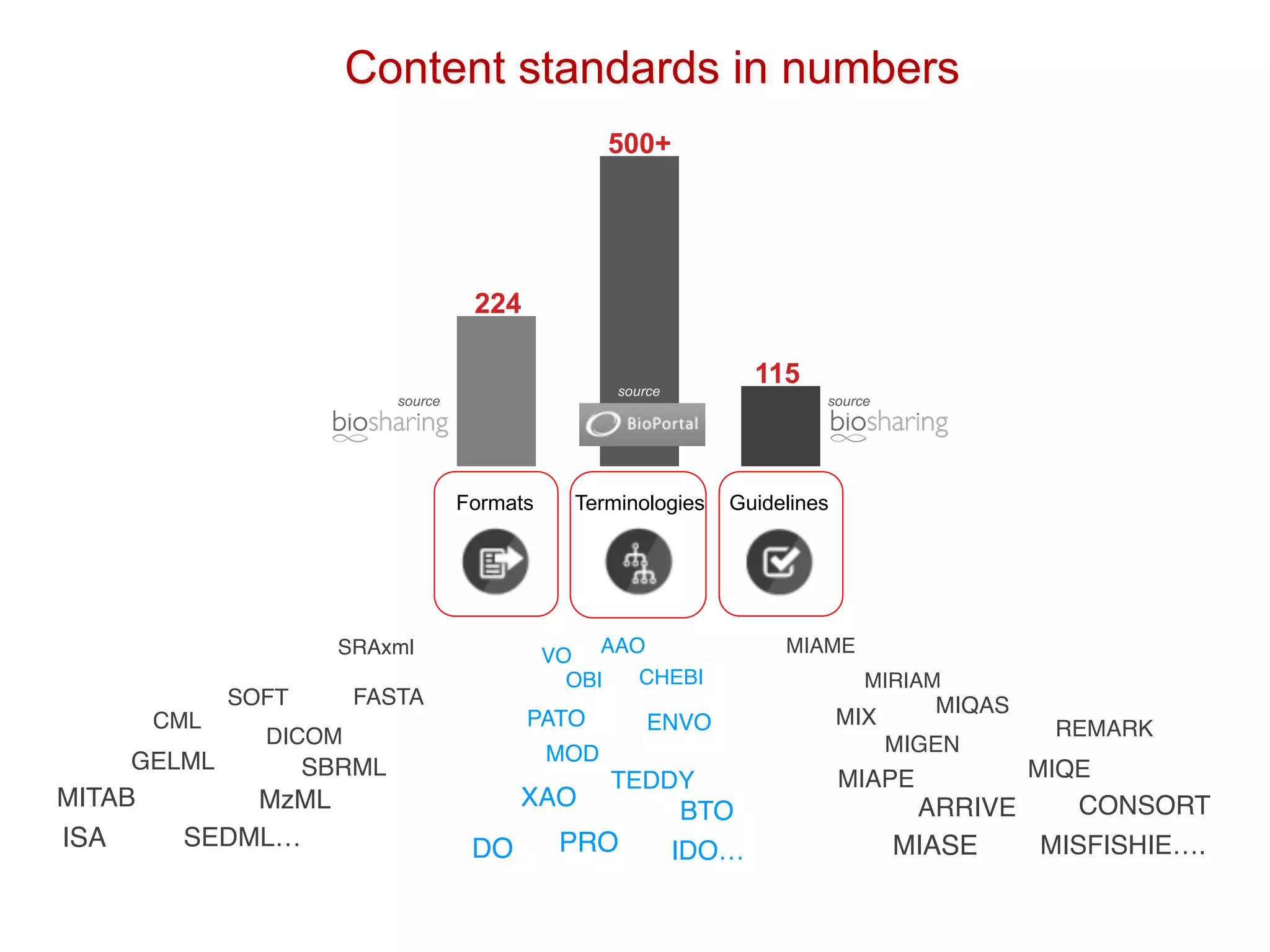
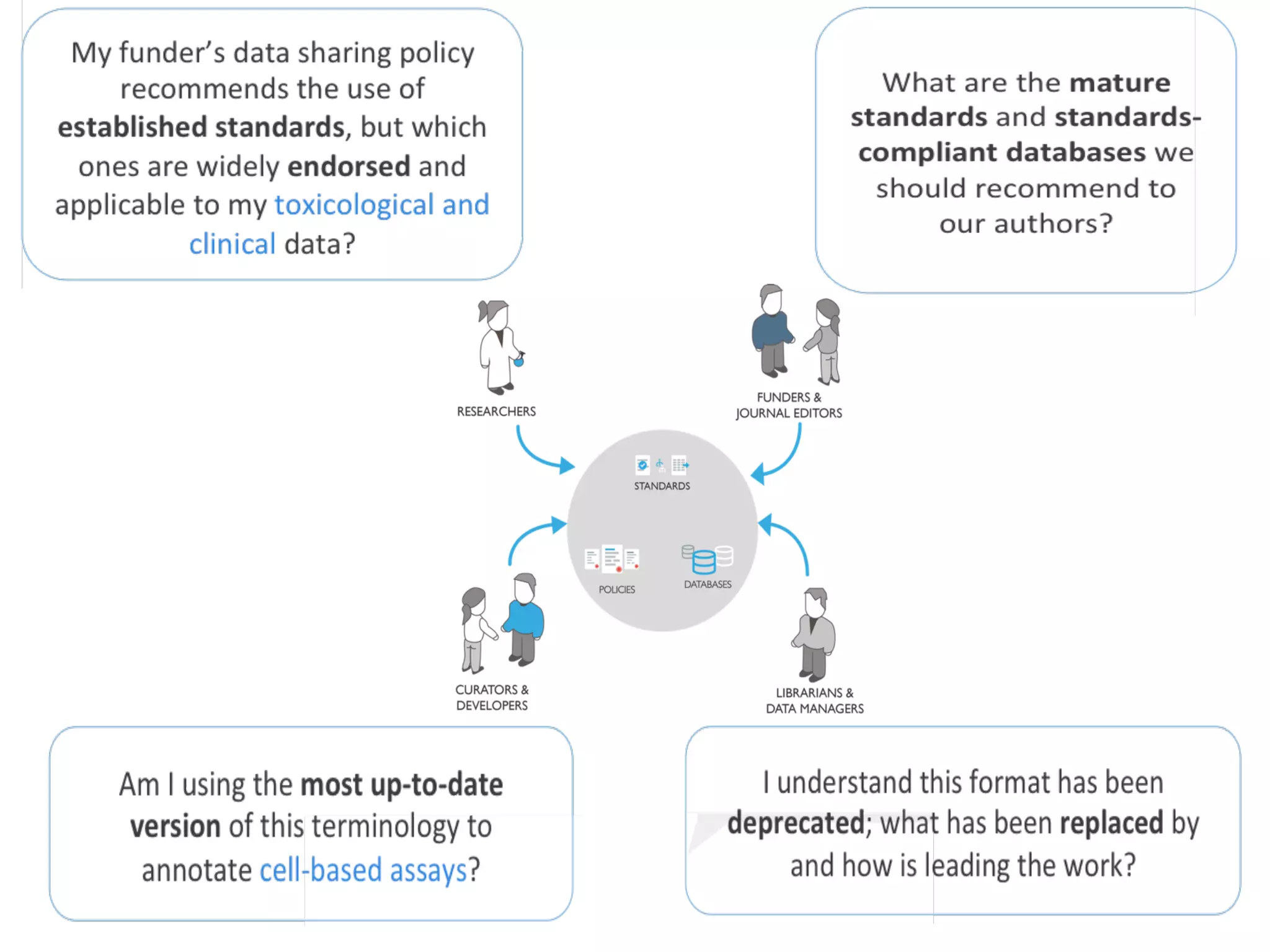
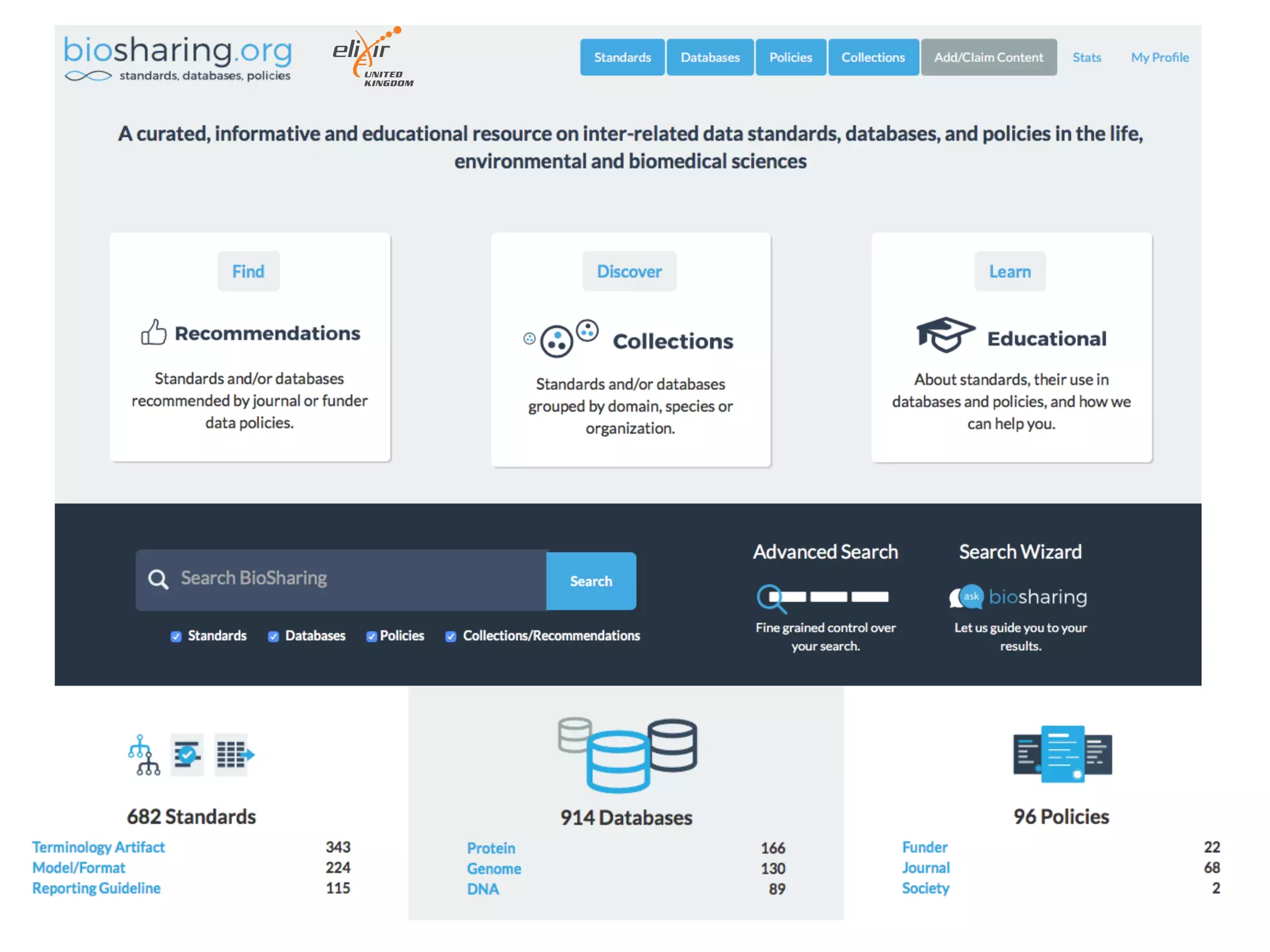
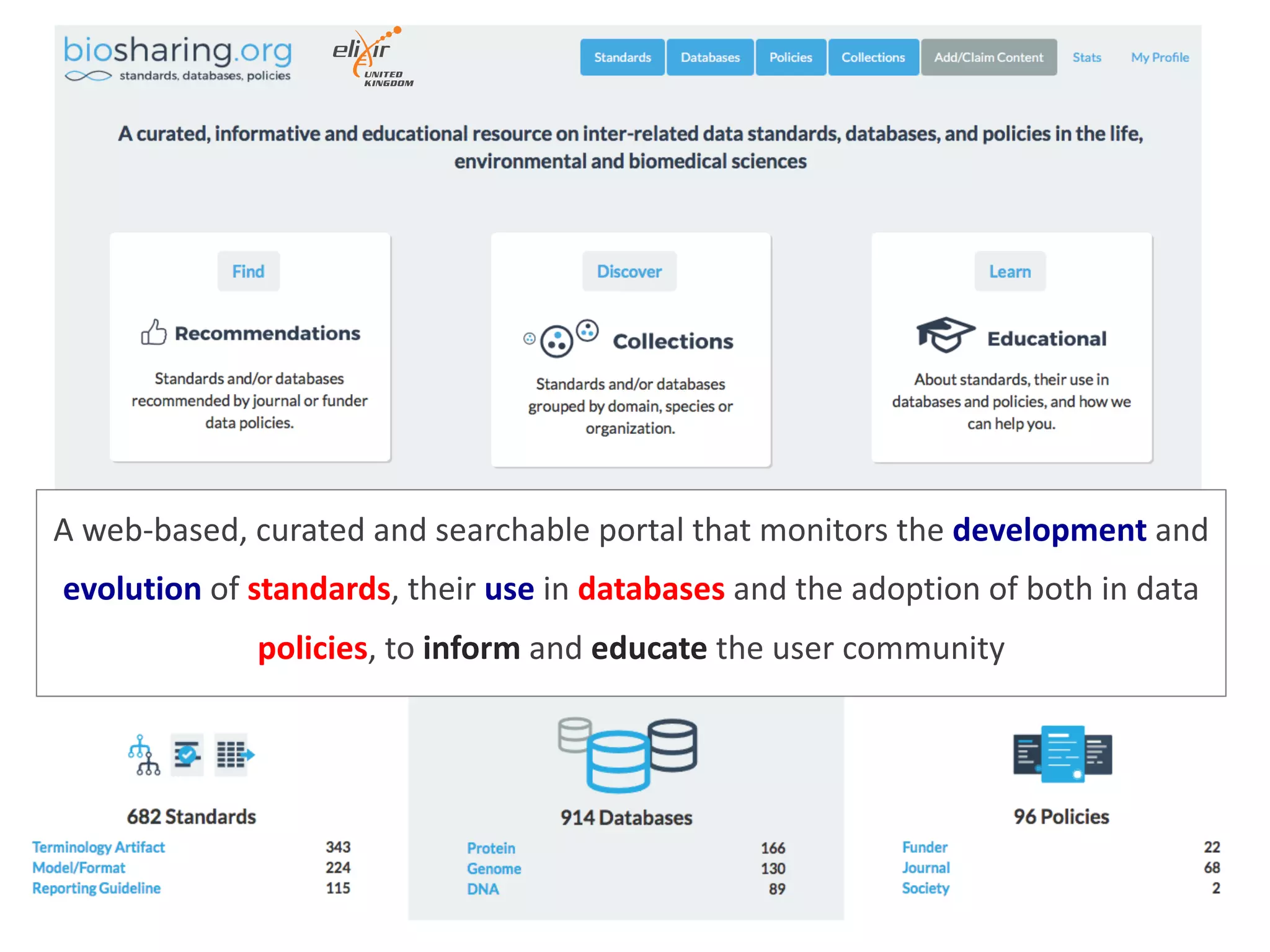
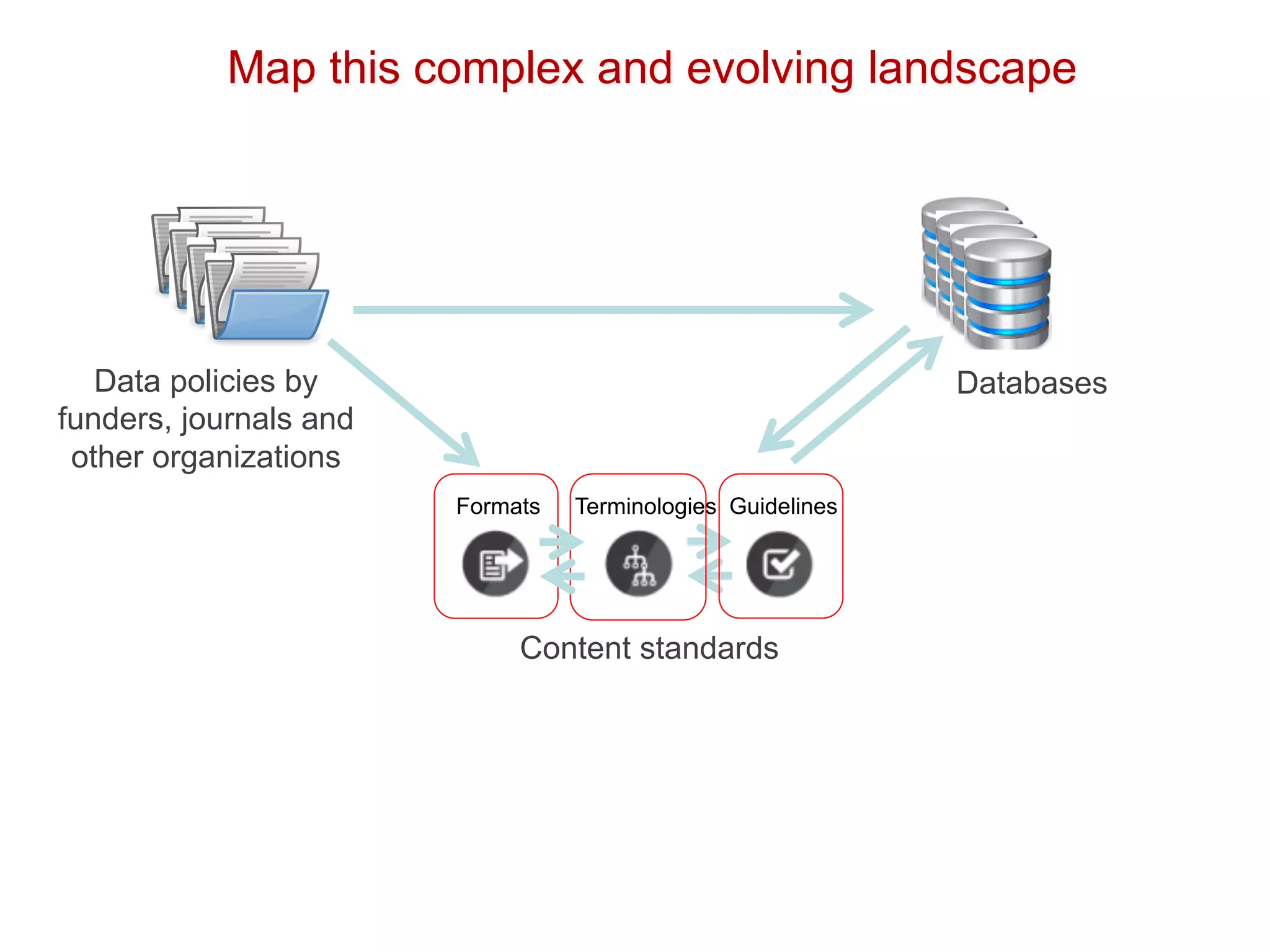
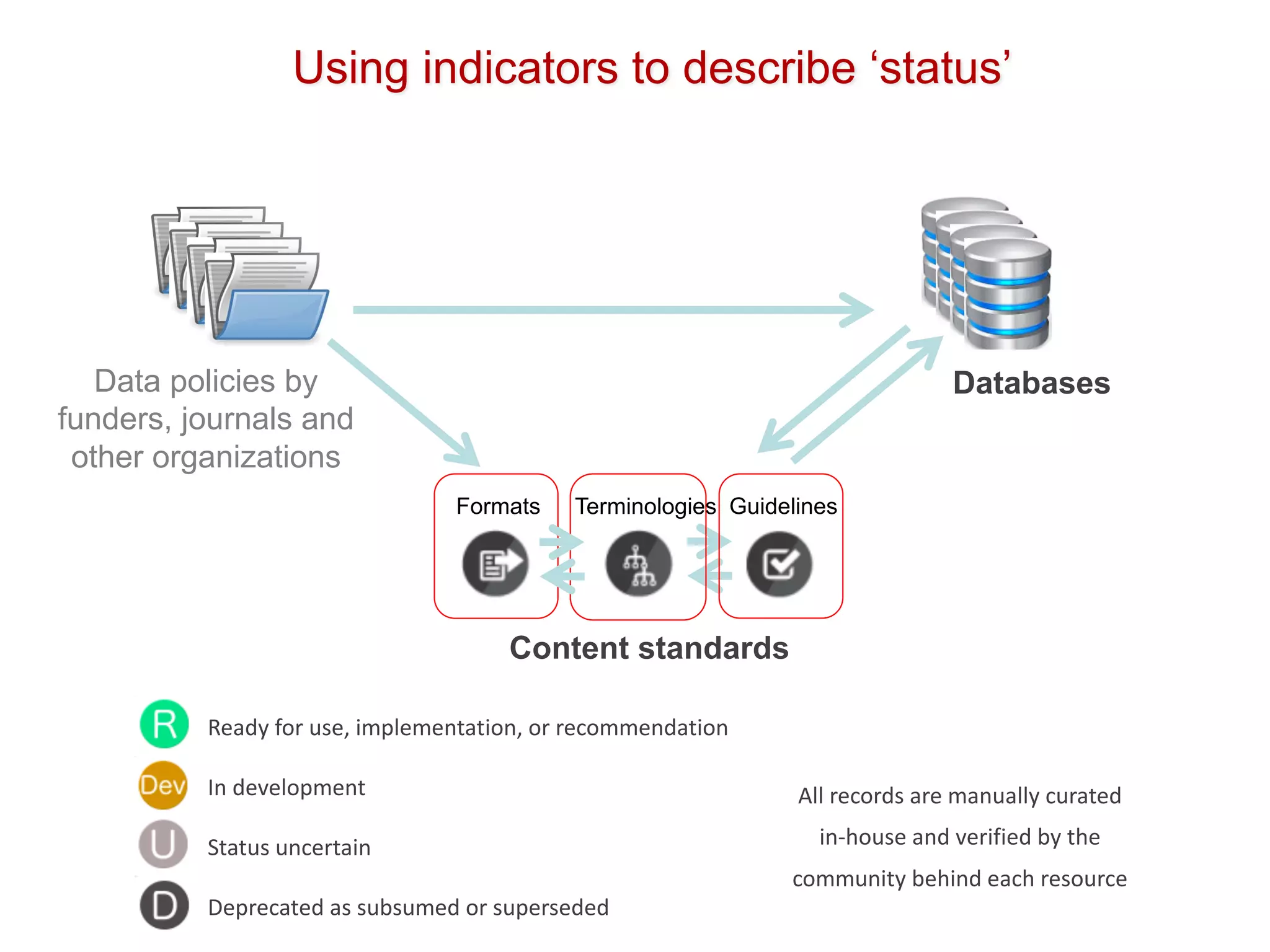
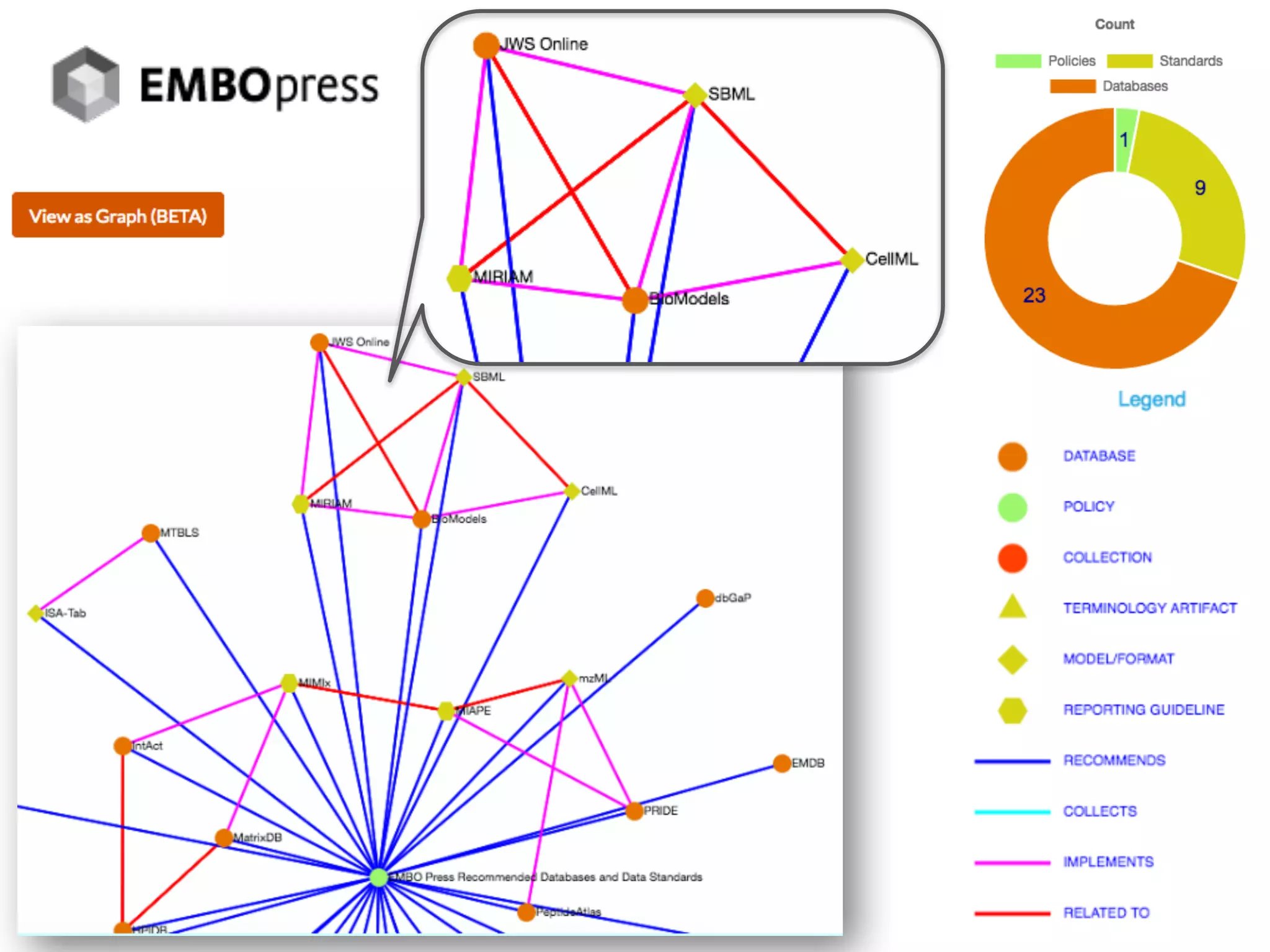
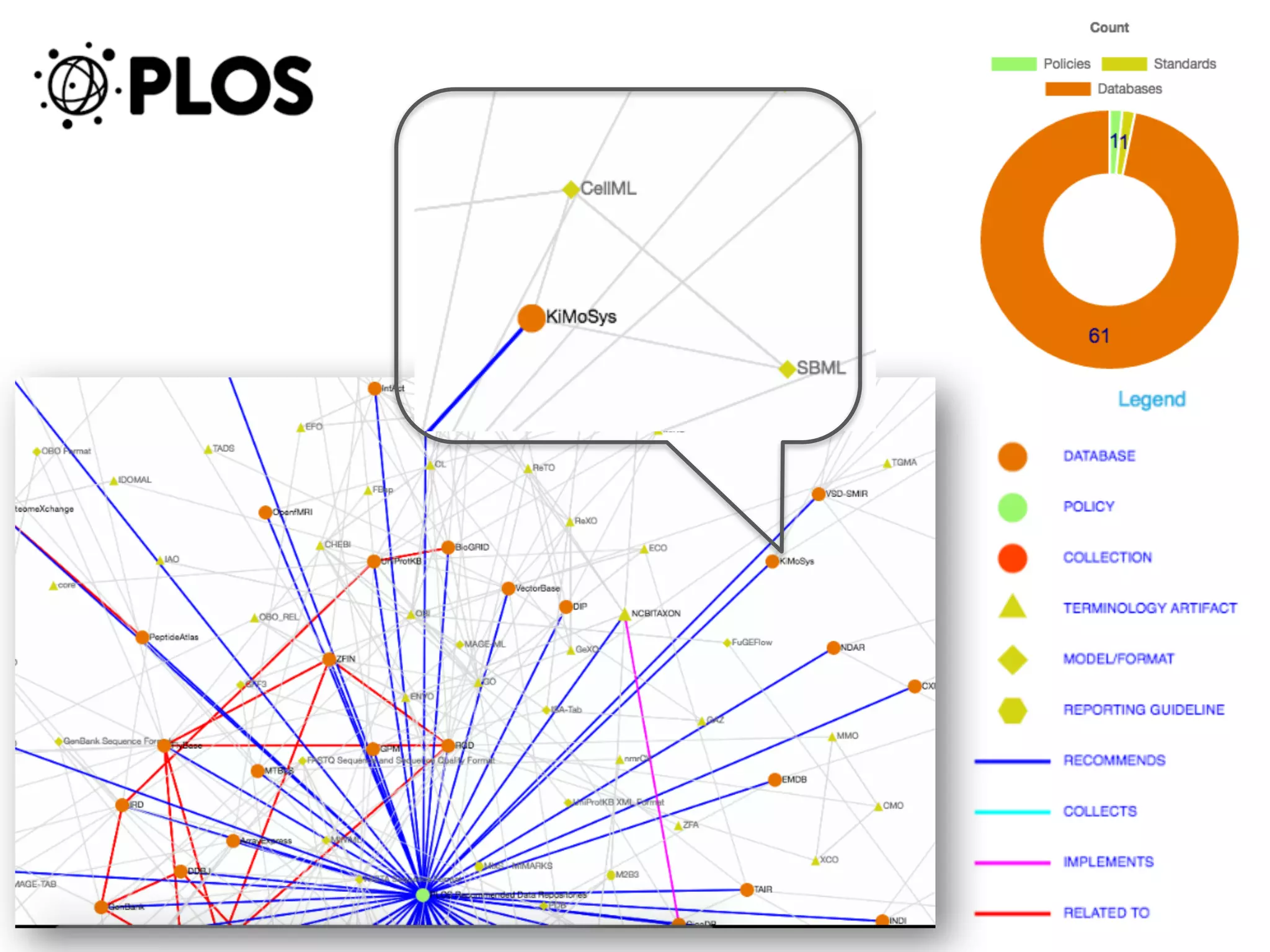
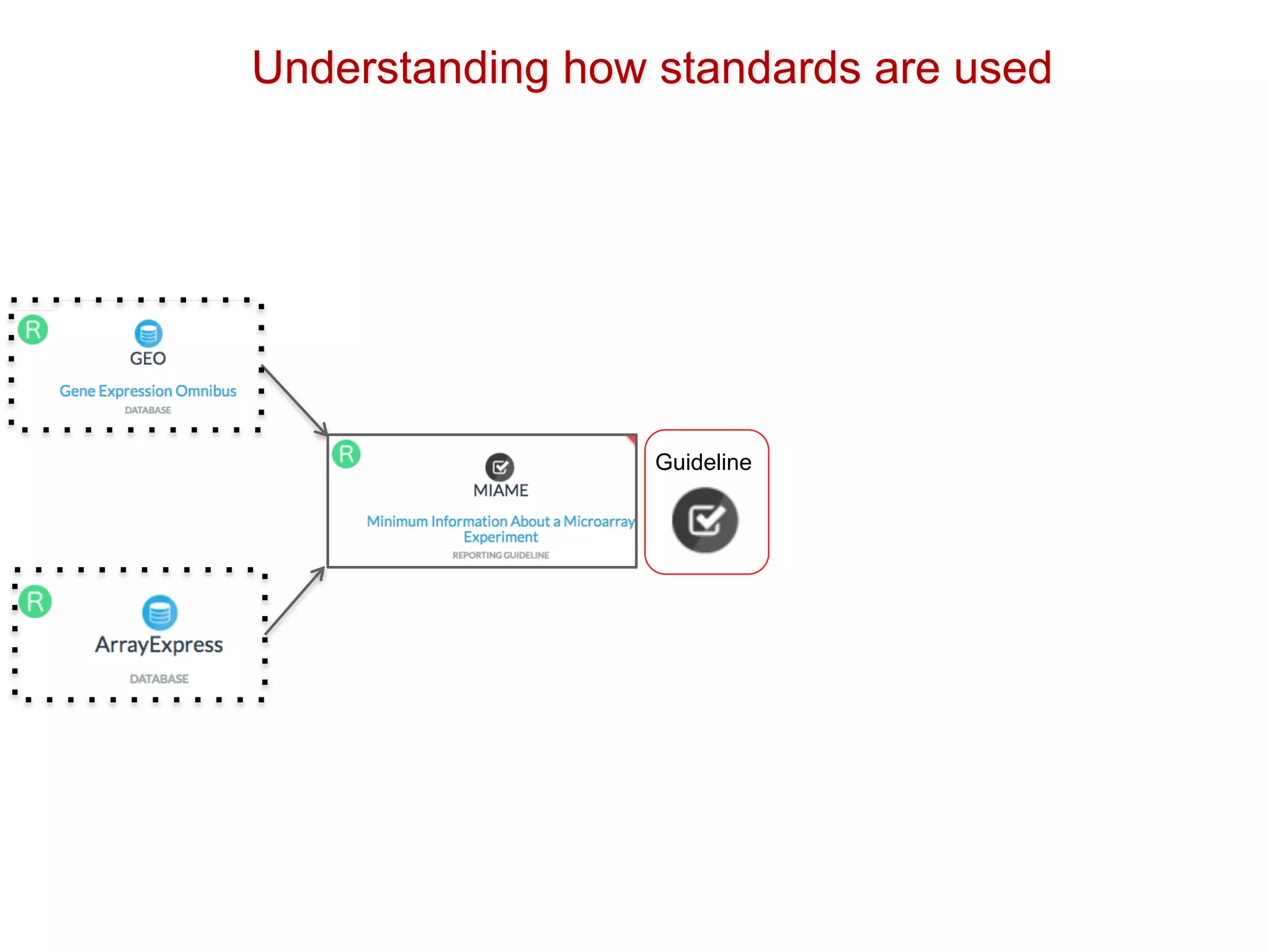
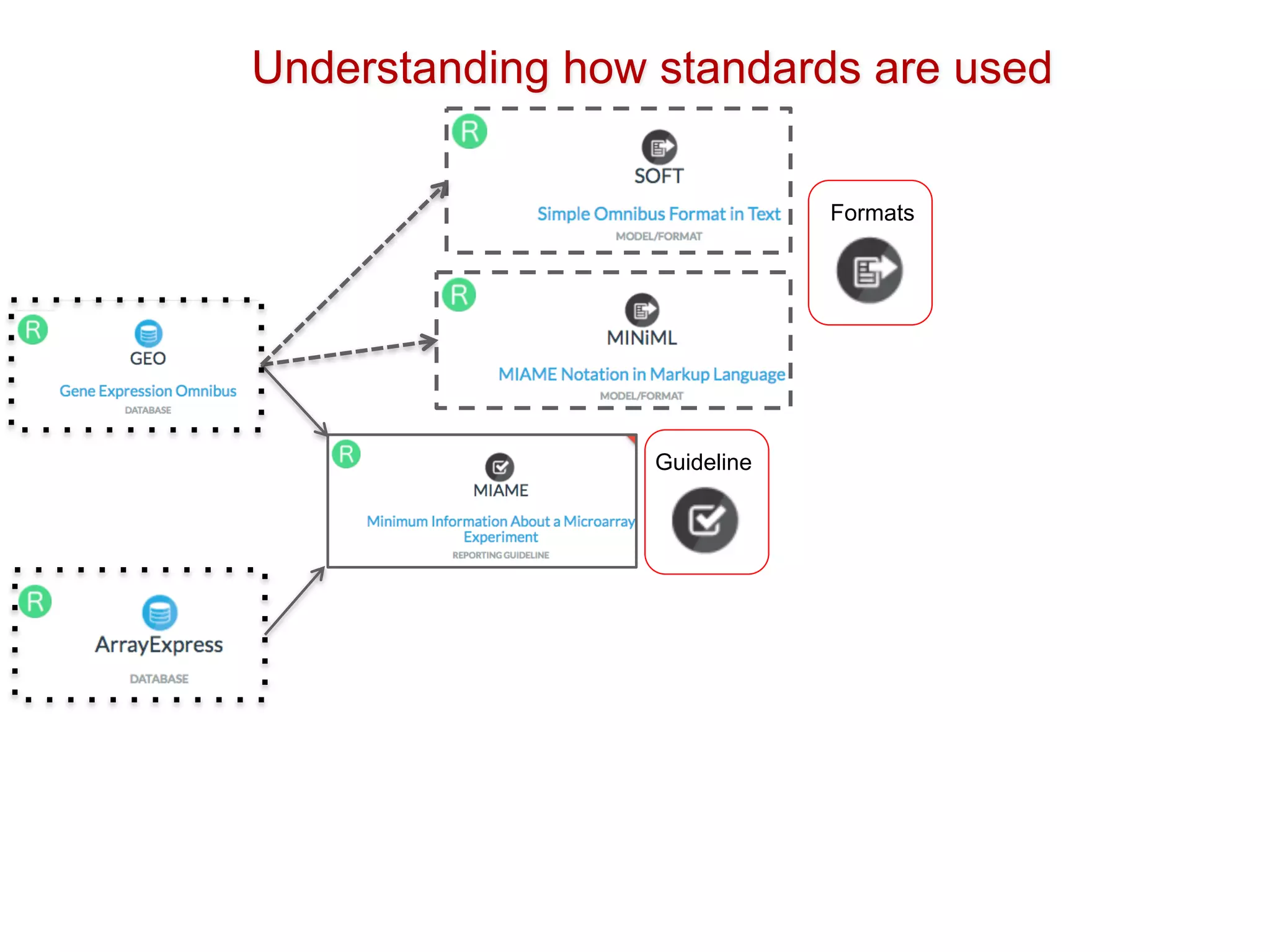
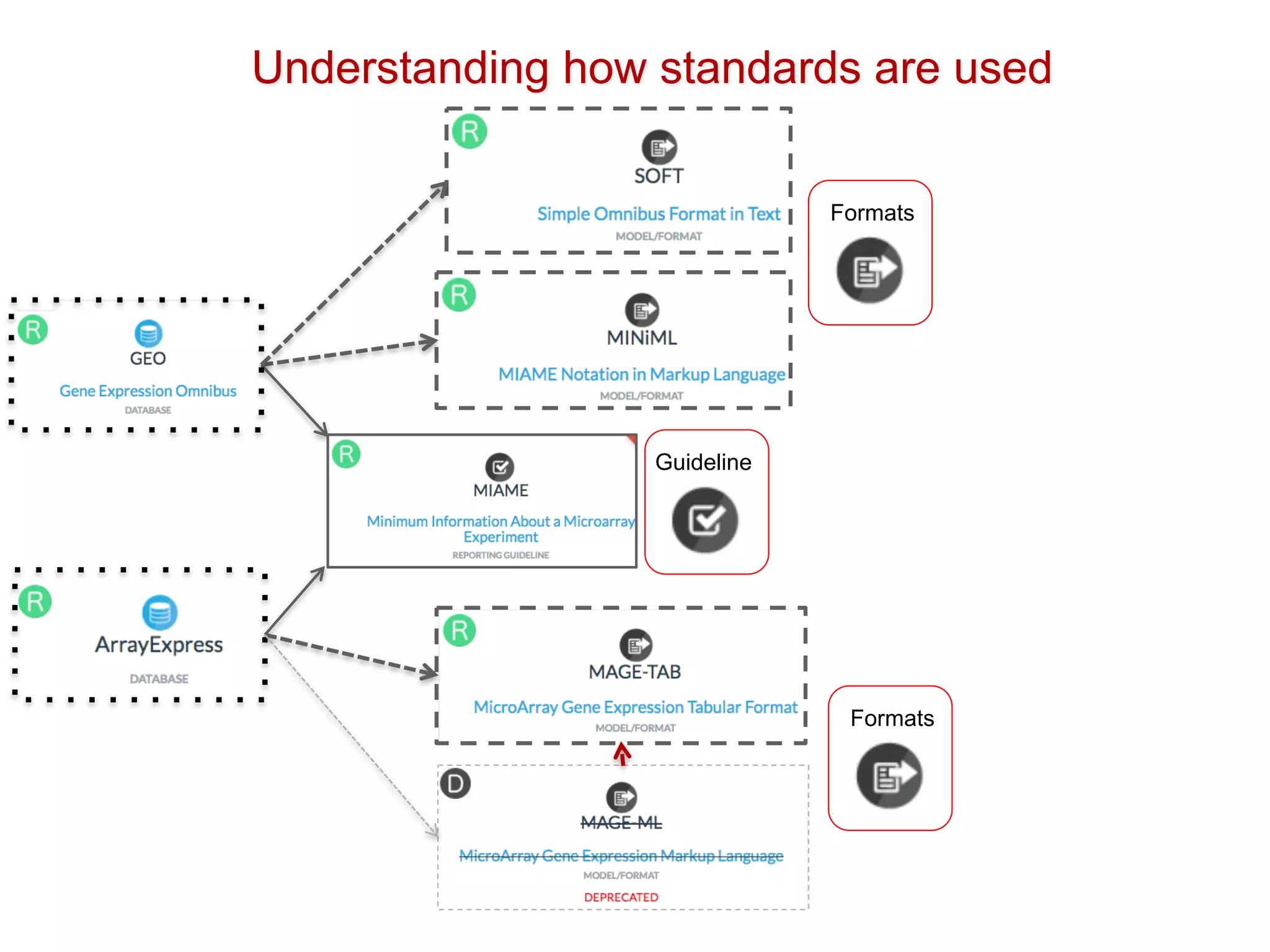
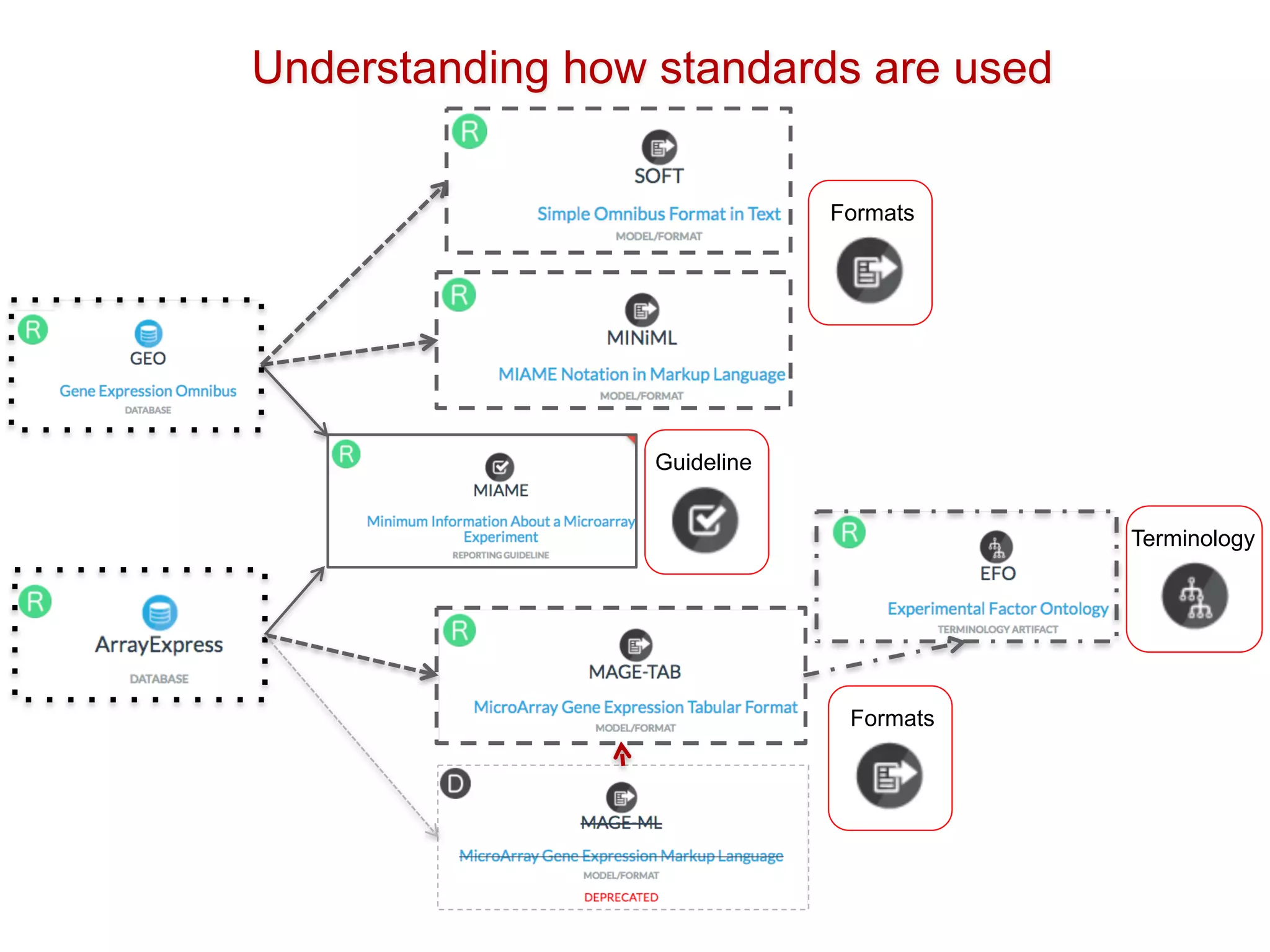
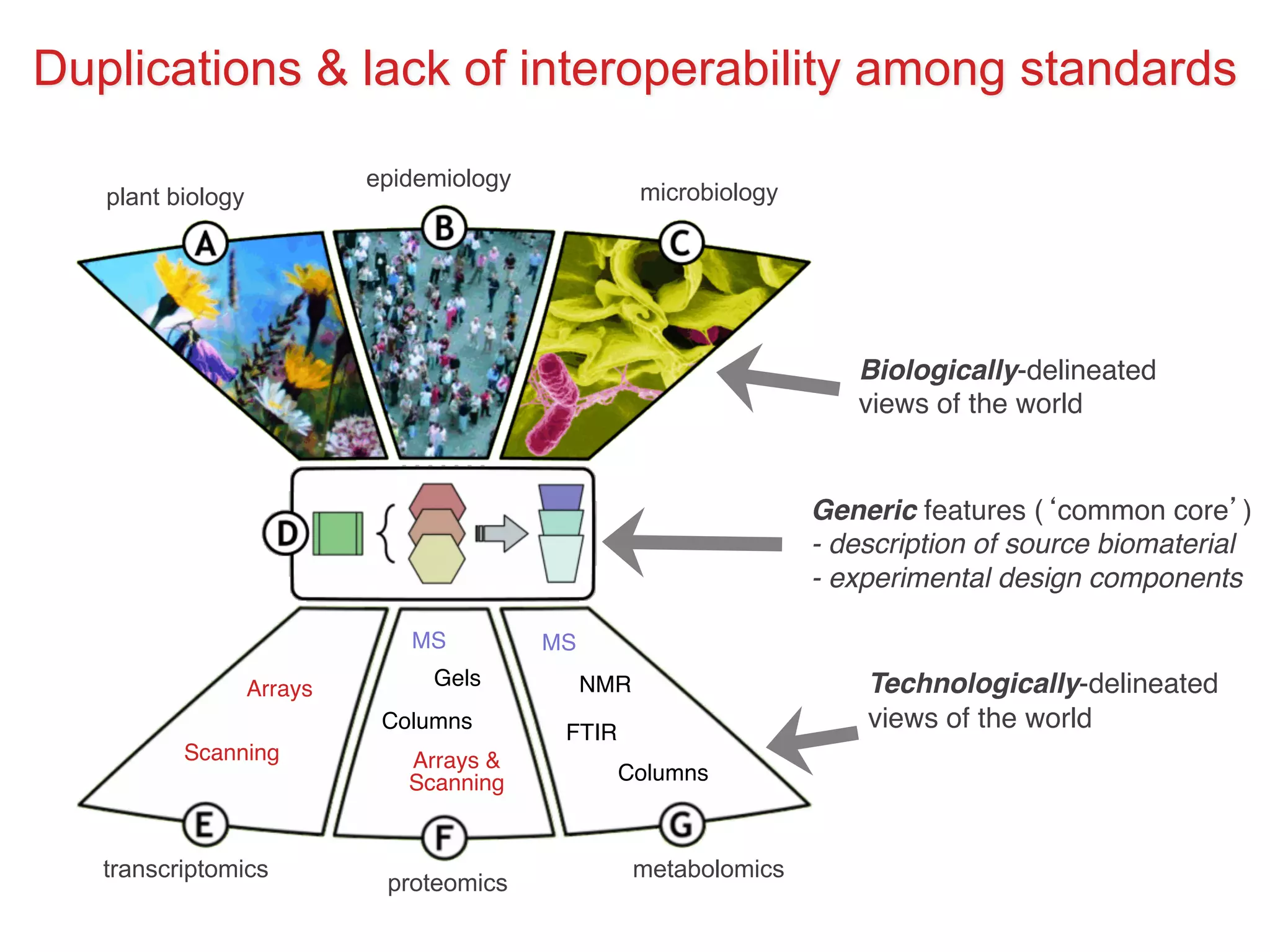
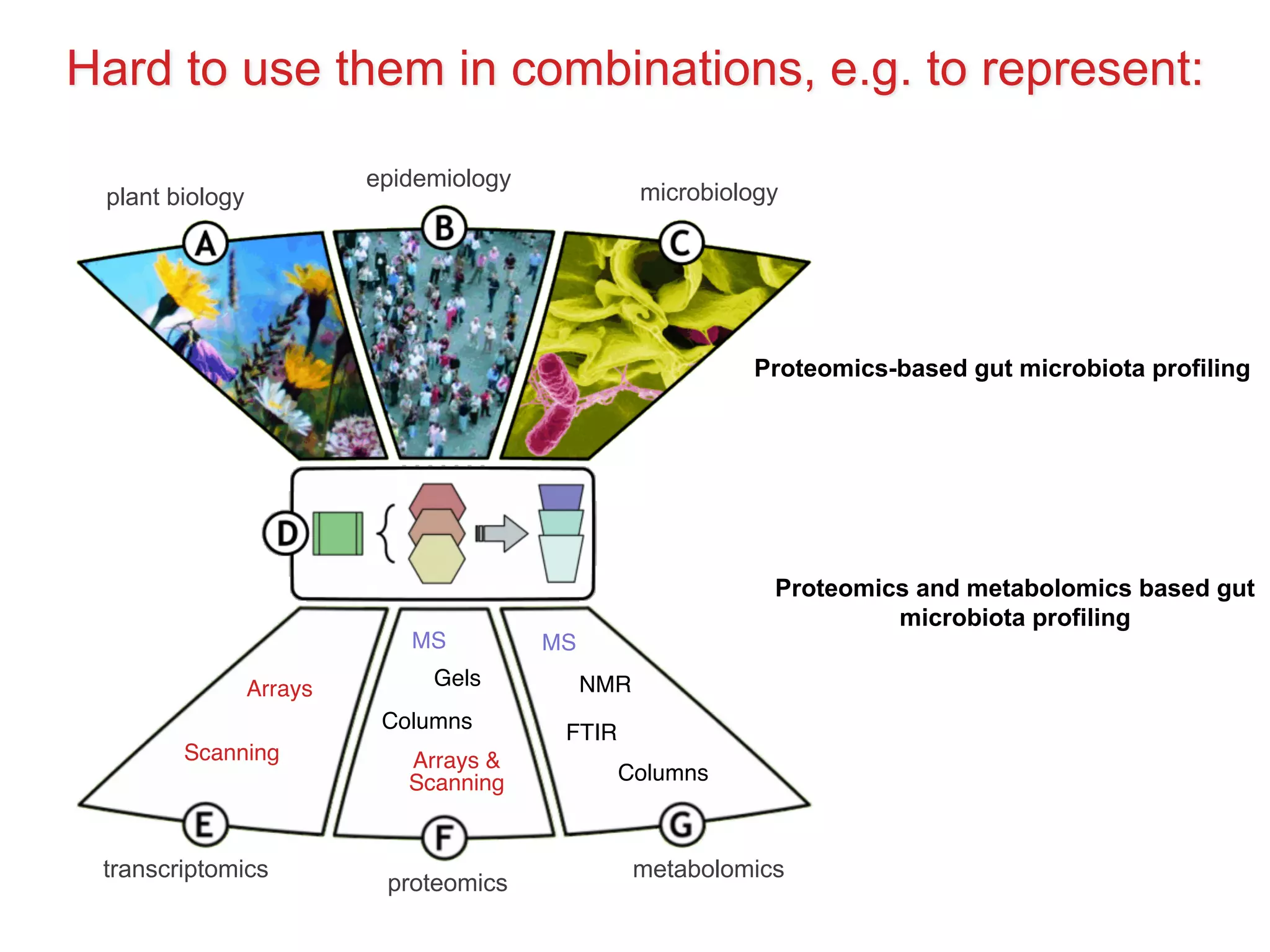
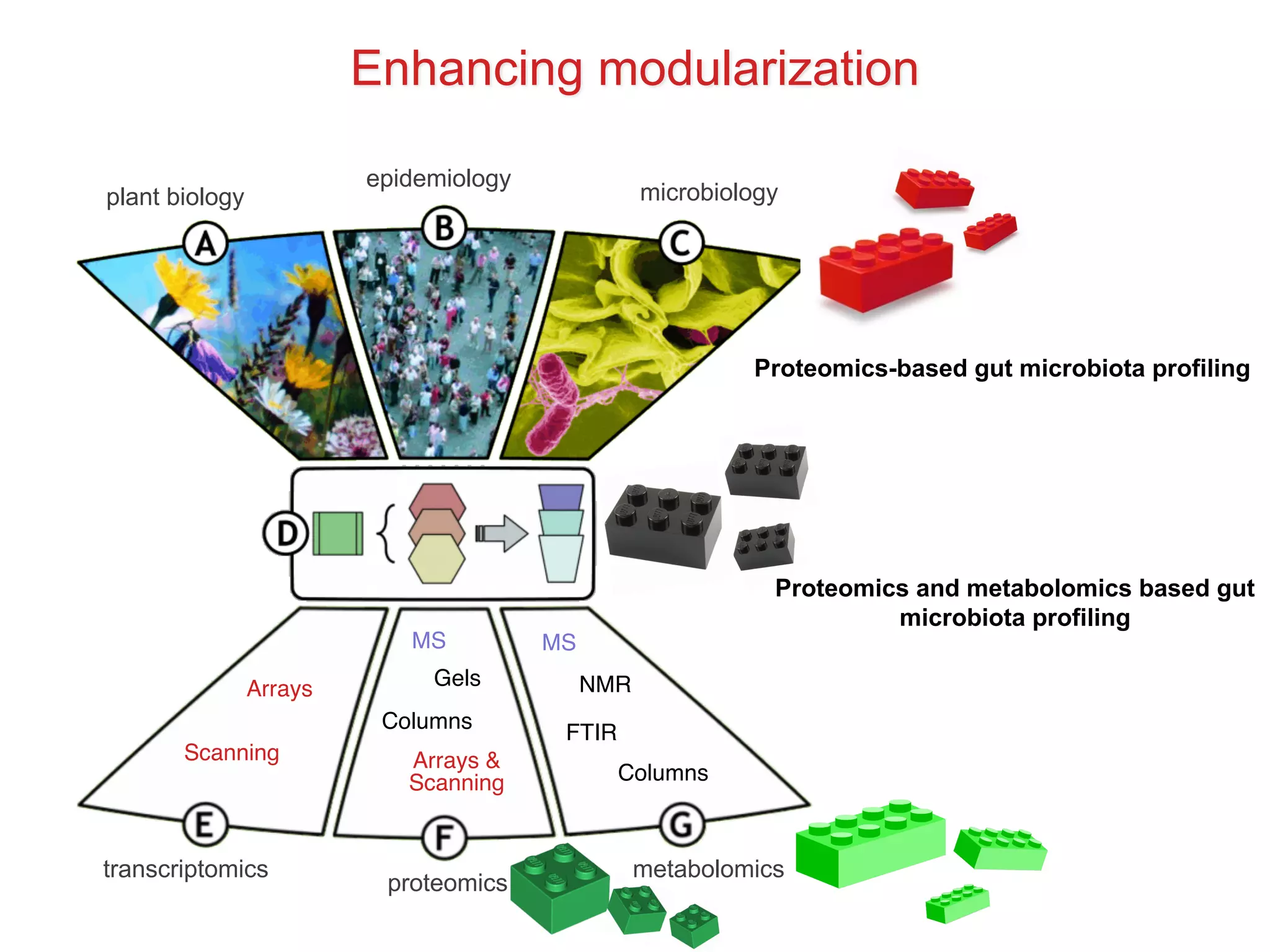
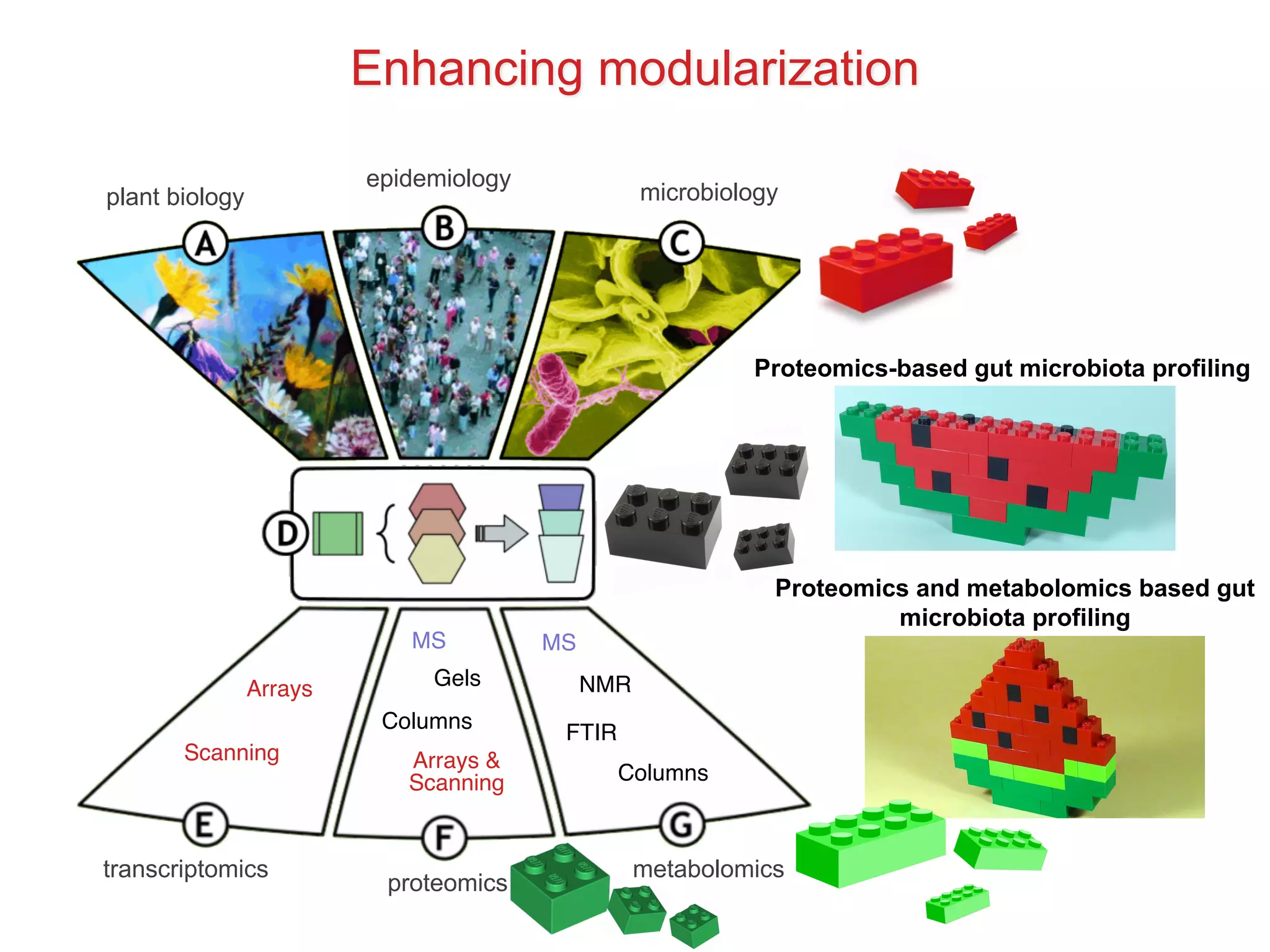
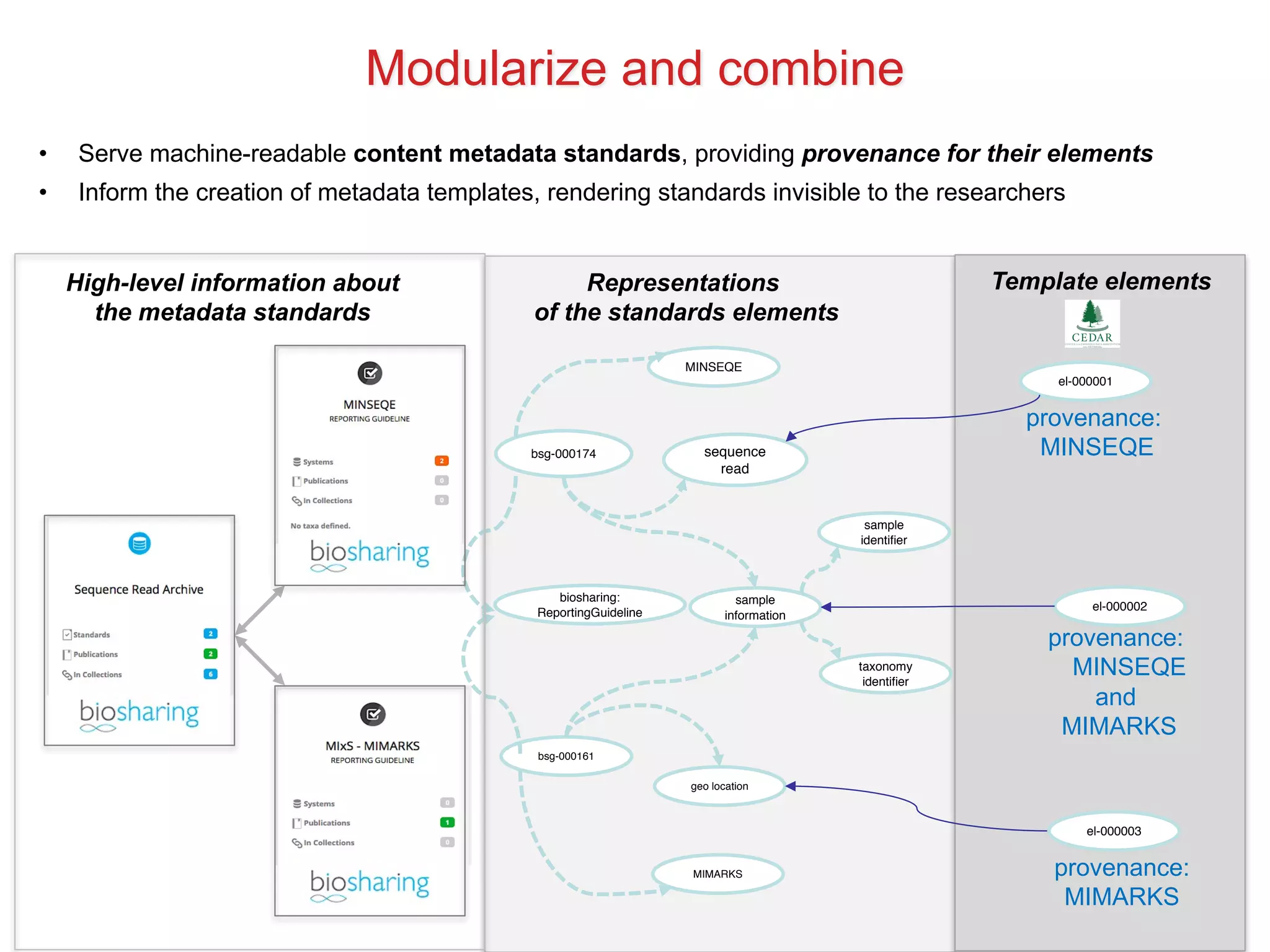
The document discusses various types of content standards including minimum reporting requirements, controlled vocabularies, and data formats that are used to ensure the interpretation, verification, and sharing of datasets. It describes efforts to map the complex landscape of evolving content standards, databases, and data policies and to understand how standards are used individually and in combination. The goal is to enhance interoperability and re-use of standards by improving modularization and representing standards elements in machine-readable templates.