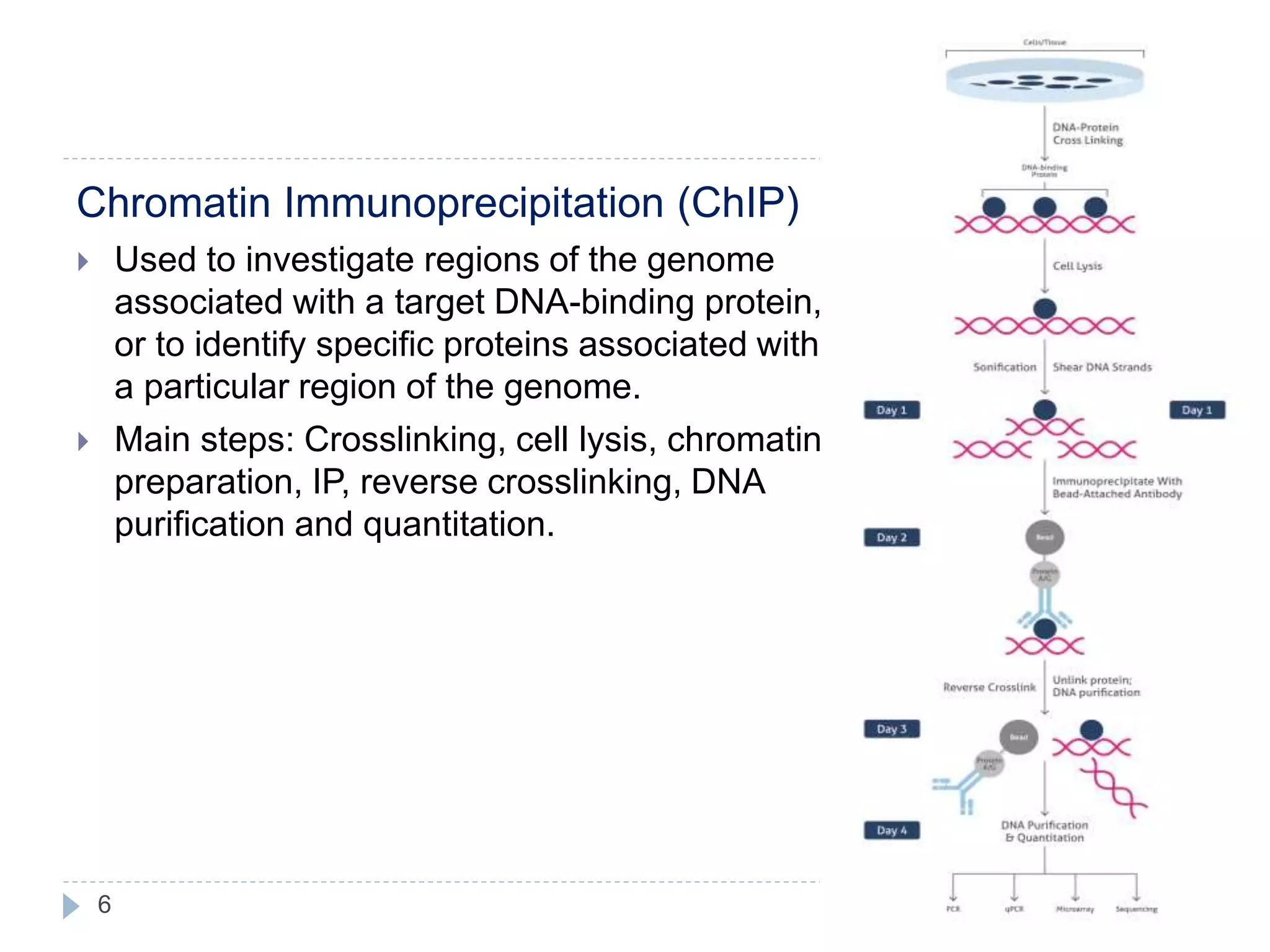
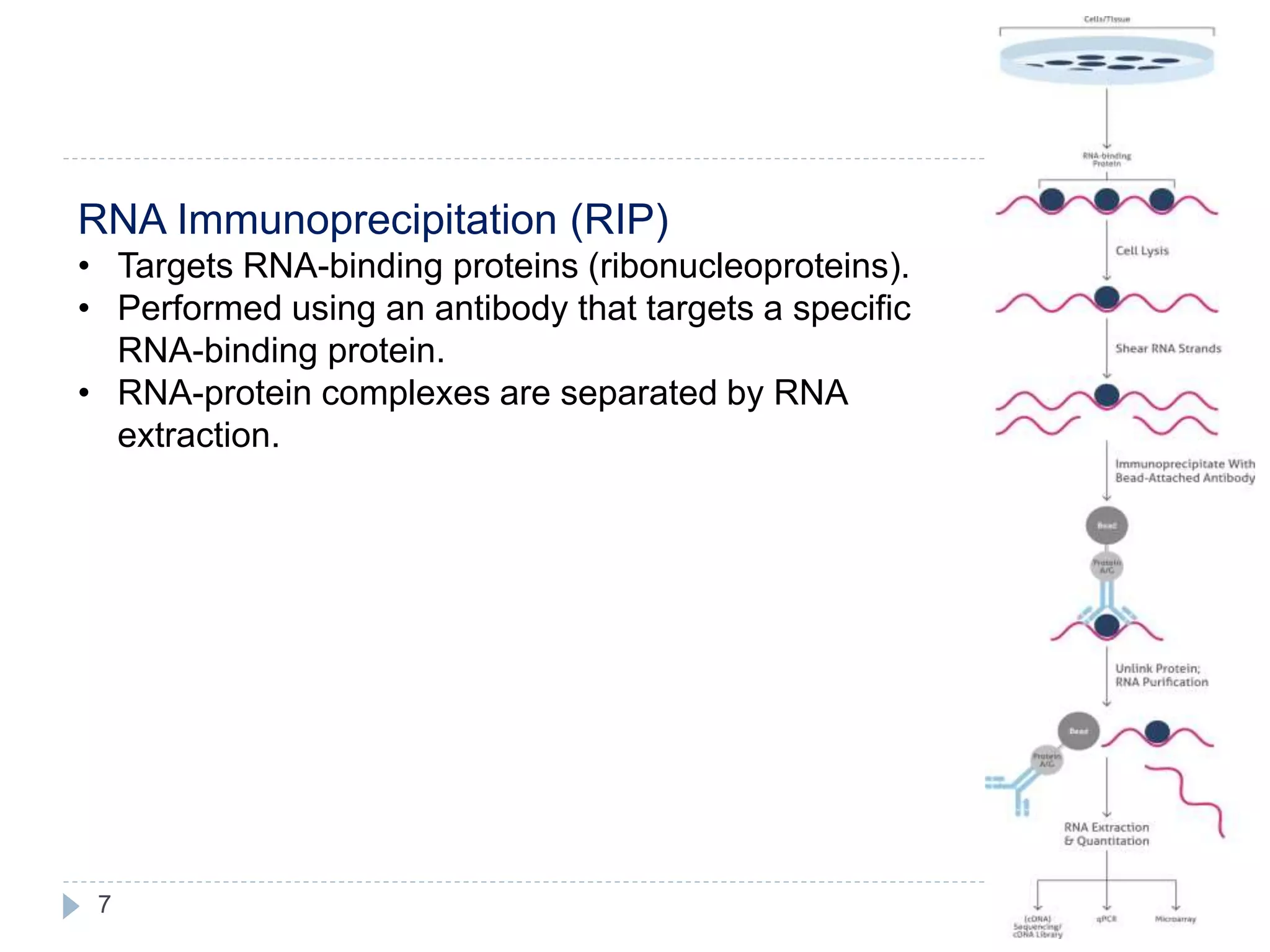
This document summarizes the technique of immunoprecipitation. It describes immunoprecipitation as a method to precipitate protein antigens out of solution using an immobilized specific antibody. The document outlines different applications of immunoprecipitation including measuring protein molecular weight, detecting post-translational modifications, and analyzing protein-protein interactions. It also describes different types of immunoprecipitation techniques such as co-immunoprecipitation, chromatin immunoprecipitation, and RNA immunoprecipitation.