HHMI Research poster -6-9-2014 Bipolar
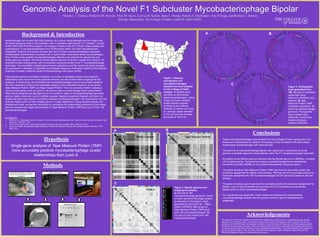
- 1. Methods • Twelve mycobacteriophages were isolated from the College of Idaho campus and 9 of these were characterized by electron microscopy. All are consistent with siphoviridae morphotypes (bacteriophages with head and tail). • The genome of mycobacteriophage Bipolar was sequenced. Comparison by whole genome nucleotide alignment places Bipolar within the F1 mycobacteriophage subcluster. • Annotation of the Bipolar genome indicates that the Bipolar genome is 58985bp, including 107 predicted genes. The genome contains a predicted programmed translational frameshift at position 9096bp (in the putative tail assembly chaperone gene). • The gene encoding Tape Measure Protein (TMP) was found to accurately predict the subcluster assignment for Bipolar. More broadly, TMP was found to accurately predict the subcluster assignment for 166 mycobacteriophages (97.5% accuracy) based on dot plot analysis. • The gene encoding Lysin A was found to accurately predict the subcluster assignment for Bipolar. Lysin A was somewhat less accurate (93.4%) in predicting the subcluster assignments for other mycobacteriophages. • Our hypothesis was supported. These results are important for understanding mycobacteriophage diversity and the genes involved in lysis of Mycobacterium smegmatis. Conclusions Genomic Analysis of the Novel F1 Subcluster Mycobacteriophage Bipolar Shandee J. Tachick, McKayla M. Stevens, Aliza M. Auces, Ljuvica R. Kolich, Hana L. Hoang, Pamela A. Dockstader, Ann P. Koga, and Richard L. Daniels Biology Department, The College of Idaho, Caldwell, Idaho 83605. Acknowledgements We would like to thank the students in the Fall BIO201 (Molecules to Cells) course for mycobacteriophage isolation, especially our TAs Macey Horch, Laura Holden, Megan Brock, Jessie Lambright and Juan Cervantes. We are very grateful to the HHMI SEA Staff, the laboratory of Graham Hatfull (University of Pittsburgh) and DNA sequencing facility at Virginia Commonwealth University. Karthik Chinnathambi and Rick Ubic provided technical assistance with electron microscopy at the Boise State University Center for Materials Characterization. The project described was supported in part by the INBRE Program, NIH Grant Nos. P20 RR016454 (National Center for Research Resources) and P20 GM103408 (National Institute of General Medical Sciences). Tape Measure Protein Figure 2. Bipolar genome and single gene analysis. A) Dot plot of 166 mycobacteriophage genomes. Colors on axes represent the phage clusters as depicted on the legend. Graph generated by Genome Pair - Rapid Dotter (GEPARD). B) Dot plot of Tape Measure Protein (TMP) gene from 166 mycobacteriophages. C) Dot plot of Lysin A gene from 166 mycobacteriophages. A Hypothesis Kalliden CuteCollette Alcor MrMud Gary1 Achille13 BipolarBactina Oink Figure 1. Electron micrographs of 9 mycobacteriophages isolated from soil samples on the College of Idaho campus. All isolates were identified as siphoviridae (tailed bacteriophages). Using ImageJ (a freely available image analysis program provided by the National Institutes of Health) we found an average capsid diameter of 73.3 nm and the average tail length of 164.8 nm. Lysin A Bacteriophages are viruses that infect bacteria. As a group, bacteriophages are the single most abundant biological entity in the biosphere, with a population estimated at 1031 particles1. As part of the HHMI SEA-PHAGES program, the College of Idaho’s Fall 2013 BIO201 class isolated and characterized 12 mycobacteriophages from Southwestern Idaho that infect Mycobacterium smegmatis. Electron microscopy revealed that each of these mycobacteriophages displayed morphological characteristics consistent with a siphoviridae morphotype (tailed bacteriophages). One of these newly-isolated mycobacteriophages (Bipolar) was selected for sequencing and further genomic analysis. We found that the Bipolar genome is 59.0kb in length and contains 107 predicted protein-coding genes, with a nucleotide sequence similar to the F1 mycobacteriophage subcluster. This annotation included gene function predictions and the results are being reviewed for submission to Genbank, a repository for biological sequence information hosted by the National Institutes of Health’s National Center for Biotechnology Information (NCBI). Following the genomic annotation of Bipolar, we further investigated whether the clustering relationships that result from whole genome similarity are also evident when single genes are analyzed. A recent study demonstrated that mycobacteriophages could be accurately assigned into their respective clusters and subclusters based on the nucleotide sequence of two genes: Tape Measure Protein (TMP) and Major Capsid Protein2. Here we examine whether ubiquitous non-structural genes, such as Lysin A, can also be used to predict phage cluster assignments. While structural genes are generally found in a similar 5’ region of mycobacteriophage genomes and are highly conserved, Lysin A exhibits a greater degree of positional freedom and does not display the same high degree of similarity between mycobacteriophages3. We hypothesize that this diversity makes Lysin A a less suitable gene for cluster assignment. Using dot plot analysis and phylogenetic trees, we test this hypothesis by visualizing the relationships predicted by both whole genome analysis and single gene analysis of Tape Measure Protein (TMP) and Lysin A from 166 mycobacteriophages. REFERENCES 1. Hatfull et al., Comparative genomic analysis of sixty mycobacteriophage genomes: Genome clustering, gene acquisition and gene size. J Mol Biol. 2010. 2. Smith et al., Phage cluster relationships identified through single gene analysis. BMC Genomics. 2013. 3. Payne and Hatfull, Mycobacteriophage Endolysins: Diverse and Modular Enzymes with Multiple Catalytic Activiites. PLOS ONE. 2012. Background & Introduction Whole Genome Singleton A B C D E F G H I J K L M O P Q R N B C Melvin(A4) Dhanush(A4) BellusTerra(A4) ICleared(A4) Flux(A4)Arturo(A4) Pukovnik(A2) SkiPole(A1) Kugel(A1) KSSJEB(A1) BPBiebs31(A1) Lesedi(A1)JC27(A1) Rosebush(B2) Qyrzula(B2) Arbiter(B2) DaVinci(A6) EricB (A6) Jeffabunny(A6) G ladiator(A6) Ham m er(A6) Blue7 (A6) Zaka (A6) HINdeR (A7) Tim shel (A7) Thibault (J) Courthouse (J) Cucu (A5) George (A5) LittleCherry (A5) Tiger (A5) RidgeCB (A1) Saintus (A8) Astro (A8) Winky (L2) Faith1 (L2) Crossroads (L2) Muddy (Singleton) Predator (H1) BigNuz (P) Konstantine (H1) Nova (D1) Troll4 (D1) Butterscotch (D1) Adjustor (D1) Gumball (D1) SirHarley (D1) Avani (F2) Jovo (A5) Toto (E)Babsiella (I1)Hedgerow (B2)Ares (B2) Send513 (R) Papyrus (R) Akoma (B3) Phyler (B3) Daisy (B3) Heathcliff (B3) Liefie (G) Hope (G)Angel (G ) G iles (Q ) Suffolk (B1) Firecracker(O ) Corndog (O) Dylan (O) BAKA (J) Optimus(J) MacnCheese(K3) Pixie(K3) Phaedrus(B3) PackMan(A9) Alma(A9) LinStu(C1) Pleione(C1) Nappy(C1) Dandelion(C1) Spud(C1)ArcherS7(C1) Ava3(C1) Brujita(I1) Island3(I1) Dori(Singleton) Butters(N) Fishburne(P)Donovan(P) Jebeks(P) LittleE(J) Phrux(E) Rakim(E) Phatbacter(E) Murphy(E) Phaux(E) Bask21(E) DS6A(Singleton)Trixie(A2) Echild(A2) L5(A2) EagleEye(A2) D29(A2) Adzzy(A2) Gadjet(B3) Bongo (M ) PegLeg (M ) IsaacEli (B1) ThreeO h3D2 (B1) Kinbote Draft (Q) UncleHowie (B1) Fang (B1) TallGrassMM (B1) Thora (B1) RockyHorror (F1) Fruitloop (F1) Shauna1 (F1) DeadP (F1) Gumbie (F1)Bipolar (F1)ChrisnMich (B4)JAMal (B4)Zemanar (B4)Nigel (B4) Stinger (B4) Cooper (B4) Anaya (K1) Angelica (K1) Adephagia (K1) JAWS (K1) BarrelRoll (K1) CrimD (K1) Validus (K1) Yoshi (F2) Redi (N) Charlie (N) ZoeJ (K2) Reprobate (B5) Acadian (B5) Twister (A10) Rebeuca (A10) Goose (A10) Rockstar (A3) HelDan (A3) Jobu08 (A3) JHC117 (A3) M icrowolf (A3) Vix (A3) M ethuselah (A3) ElTiger69 (A5) Benedict(A5) SG 4 (F1) Bernardo (B3) Ramsey(F1) Jabbawokkie(F2) TM4(K2) Rey(M) Patience(Singleton) Rumpelstiltskin(L2) UPIE(L1) JoeDirt(L1)LeBron(L1) Single-gene analysis of Tape Measure Protein (TMP) more accurately predicts mycobacteriophage cluster relationships than Lysin A. A Figure 3. Phylogenetic trees generated from 166 A) Lysin A nucleotide sequences using Maximum Likelihood (ML) analysis. B) Tape Measure Protein (TMP) nucleotide sequences. C) whole mycobacteriophage genomes. Phylogenetic trees created using Molecular Evolutionary Genetics Analysis software (MEGA6). Adephagia(K1) JAWS(K1) BarrelRoll(K1) Anaya(K1) Angelica(K1) Pixie(K3) EagleEye(A2) Jabbawokkie(F2) Alma(A9) Courthouse(J) Liefie(G) Redi(N) Bongo(M)Fang(B1) Twister(A10)D29(A2) SkiPole(A1) Rosebush(B2) Ares(B2) Hedgerow (B2) Arbiter(B2) Q yrzula (B2) Validus (K1) Akom a (B3) Heathcliff (B3) Bernardo (B3) Gadjet (B3) Phlyer (B3) Daisy (B3) Cuco (A5) George (A5) Tiger (A5) Phaedrus (B3) Brujita (I1) Island3 (I1) HINdeR (A7) Timshel (A7) Suffolk(B1) ThreeOh3D2 (B1) IsaacEli (B1) Thora (B1) TallGrassMM (B1) UncleHowie (B1) Astro (A8) Saintus (A8) ElTiger69 (A5) Phelemich (B5) Rockstar (A3) HelDan (A3)PackMan (A9)Pukovnik (A2)BellusTerra (A4) TiroTheta9 (A4)Flux (A4)Melvin (A4) ICleared (A4) Arturo (A4) Methuselah (A3) Trixie (A2) Goose (A10) Rebeuca (A10) Reprobate (B5) Kayacho (B4) Rey (M ) Butters (N) C harlie (N) C orndog (O ) BPs (G ) Hope (G ) Avrafan (G) Angel(G) Phrux(E) Phatbacter(E) Phaux(E) Rakim (E) Bask21(E) Murphy(E) Toto(E) Bipolar(F1)Shauna1(F1) DeadP(F1)SG4(F1) Fruitloop(F1) GUmbie(F1) Ramsey(F1) RockyHorror(F1) DS6A(Singleton) Avani(F2) Yoshi(F2) Giles(Q) Kinbote(Q)HH92(Q) JHC117(A3) Vix(A3) Jobu08(A3) Microwolf(A3) PegLeg(M) TM4(K2) Echild(A2) CrimD(K1) MacnCheese(K3) JAMal(B4) Zemanar(B4) ArcherS7(C1) Dandelion(C1) Spud(C1) Nappy(C1) Ava3 (C1) Pleione (C1) N igel(B4) Stinger (B4) Acadian (B5) Dylan (O) Firecracker (O) ZoeJ (K2) Blue7 (A6) Hammer (A6) Gladiator (A6) DaVinci (A6) EricB (A6)L5 (A2)LinStu (C1)Kugel (A1)Lesedi (A1)Dhanush (A4)JC27 (A1)RidgeCB (A1)Donovan (P) Jebeks (P) Fishburne (P) BigNuz (P) LittleE (J) BAKA (J) Optimus (J) Thibault (J) ChrisnMich (B4) Dori (Singleton) LittleCherry (A5) Jovo (A5) Jeffabunny (A6) Zaka (A6) BPBiebs31 (A1) KSSJEB (A1) Benedict (A5) Adzzy (A2) Babsiella (I1) Cooper (B4) Predator (H1) Gumball (D1) Nova (D1) SirHarley (D1) Troll4 (D1) Adjutor (D1) Butterscotch (D 1) Konstantine (H1) Papyrus (R) Send513 (R) Patience(Singleton) Faith1(L2) W inky(L2)Crossroads(L2) Rumpelstiltskin(L2) Muddy(Singleton) UPIE(L1) JoeDirt(L1)LeBron(L1) C B Phatbacter(E) Phaux(E)Murphy(E) Bask21(E) Phrux(E) Rakim(E) Toto(E) BigNuz(P)Donovan(P) Fishburne(P) Jebeks(P) Rebeuca(A10)Twister(A10)Goose(A10) Microwolf(A3) Jobu08(A3) JHC117(A3)Vix(A3) Methuselah (A3) ElTiger69 (A5) Benedict(A5) LeBron (L1) U PIE (L1) JoeDirt (L1) Rum pelstiltskin (L2) Faith1 (L2) W inky (L2) Crossroads (L2) Arturo (A4) Flux (A4) ICleared (A4) Adzzy (A2) Pukovnik (A2) EagleEye (A2) Tiger (A5) Jovo (A5) Cuco (A5) George (A5) LittleCherry (A5) ArcherS7 (C1) Ava3 (C1) Pleione (C1) Nappy (C1) Dandelion(C1) TiroTheta9 (A4) Dhanush (A4) BellusTerra (A4) Melvin (A4) HINdeR (A7)Timshel (A7)Trixie (A2)Echild (A2)L5 (A2)D29 (A2)DaVinci (A6)EricB (A6)Zaka (A6) Jeffabunny (A6) Gladiator (A6) Blue7 (A6) Hammer (A6) Yoshi (F2) Avani (F2) Jabbawokkie (F2) Predator (H1) Konstantine (H 1) Lesedi(A1) BPBiebs31 (A1) SkiPole (A1) JC27 (A1) KSSJEB (A1) Kugel(A1) RidgeCB(A1) Brujita(I1) Island3(I1) Babsiella(I1) TM4(K2) ZoeJ(K2) DS6A(Singleton) TallGRassMM(B1) Firecracker(O) Corndog(O) Dylan(O) KinboteDraft(Q) Giles(Q) HH92(Q) Nova(D1) Troll4(D1) Adjutor(D1) Butterscotch(D1)SirHarley(D1)Gumball(D1) Patience(Singleton) PackMan(A9)Alma(A9) Rockstar(A3) HelDan(A3) Saintus(A8) Astro(A8) LinStu(C1) Spud(C1) LittleE(J) Courthouse(J) Thibault(J) BAKA (J) Optimus(J) Send513 (R) PAPYRUS (R) M uddy (Singleton) R ey (M ) Bonga (M ) PegLeg (M ) Phlyer (B3) Phaedrus (B3) Heathcliff (B3) Akoma (B3) Bernardo (B3) Gadjet (B3) Daisy (B3) Charlie (N)Redi (N)Butters (N)Validus (K1)CrimD (K1)BPs (G) Avrafan (G)Hope (G) Angel (G) Liefie (G) MacnCheese (K3) Pixie (K3) Anaya (K1) Adephagia (K1) Angelica (K1) JAWS (K1) BarrelRoll (K1) Fang (B1) ThreeOh3D2 (B1) Thora (B1) IsaacEli (B1) UncleHowie (B1) Suffolk (B1) Cooper (B4) Stinger (B4) Nigel (B4) JAMal (B4) ChrisnMich (B4) Zemanar (B4) Reprobate (B5) Phelem ich (B5) Acadian (B5) Kayacho (B4) Ares (B2) R osebush (B2) Hedgerow (B2) Q yrzula (B2) Arbiter(B2) Dori(Singleton) Ramsey(F1) RockyHorror(F1) DeadP(F1) SG4(F1) Shauna1(F1)Fruitloop(F1) Bipolar(F1)GUmbie(F1)