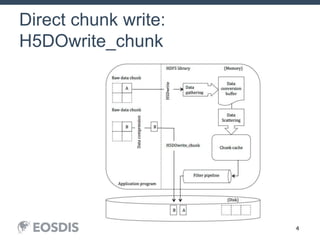
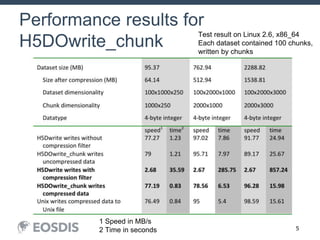
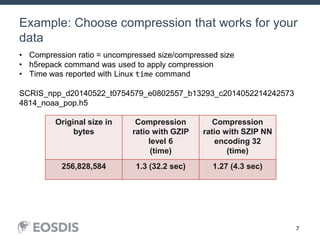
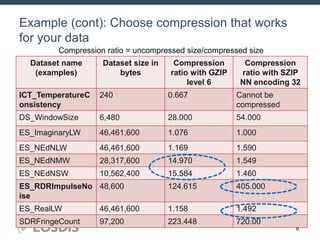
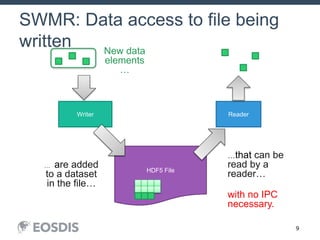
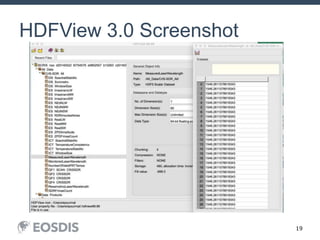
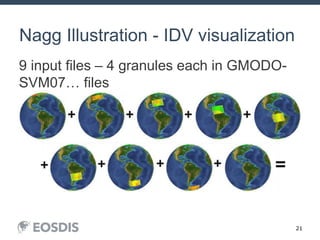
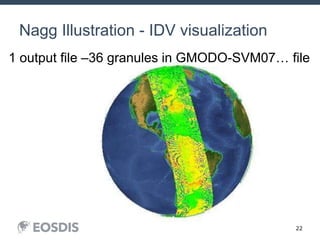
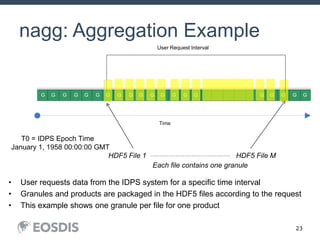
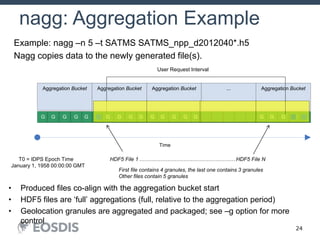
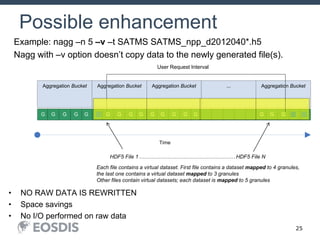
The HDF Group provides updates on new features in HDF including faster compression, single writer/multiple reader file access, virtual datasets, and dynamically loaded filters. They also discuss tools like HDFView, nagg for data aggregation, and a new HDF5 ODBC driver. The work is supported by NASA.