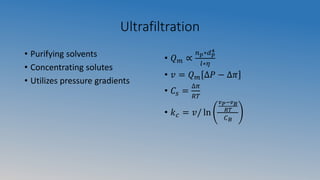
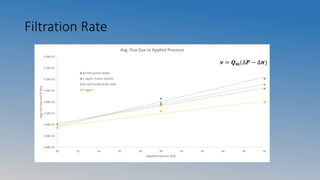
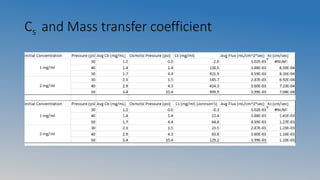
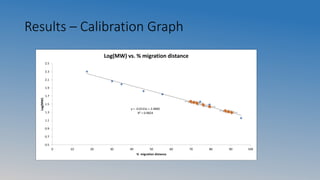
This document summarizes an experiment on ultrafiltration and gel electrophoresis. For ultrafiltration, the goals were to understand membrane separation principles and parameters like osmotic pressure and fouling. Experiments tested filtration rate with different protein concentrations. Results showed osmotic pressure had a greater effect on mass transfer than fouling. For gel electrophoresis, the goals were to separate and measure molecular weights of pepsin and rennet using SDS-PAGE. Results showed crude pepsin, pure pepsin, and rennet-pepsin mixture were close to literature values, while rennet alone had some unexpected bands possibly from breakdown.