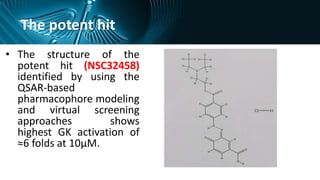
This document discusses using computer-aided drug design (CADD) to develop new treatments for type 2 diabetes by targeting glucokinase activators. It describes using pharmacophore modeling, virtual screening, and molecular docking to identify potential drug candidates from large databases. Specifically, it details a case study where these CADD methods were used to discover a novel glucokinase activator that showed 6.3-fold activation at 10 μM in in vitro bioassays. The potent hit compound identified may help control blood glucose levels and improve outcomes for patients with type 2 diabetes.