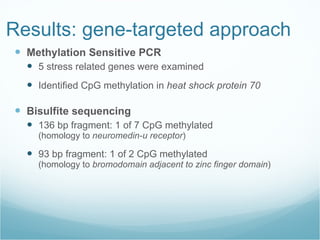
The document summarizes research on DNA methylation patterns and epigenetic regulation in the Pacific oyster. Results showed DNA methylation occurs in stress-related genes and may play a regulatory role in immune and stress responses. Understanding genetic and epigenetic influences could increase predictability in selective breeding of oysters. Further evaluating epigenetic mechanisms in bivalves using new methods like whole genome bisulfite sequencing may provide insights with implications for selective breeding, hybrid vigor, and how nutrition affects gene expression through DNA methylation.