Ivan Sotelo Poster FINAL Ver
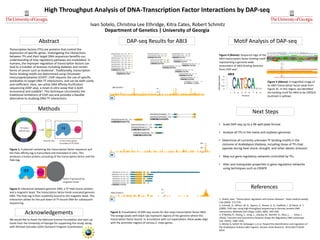
- 1. High Throughput Analysis of DNA-Transcription Factor Interactions by DAP-seq Ivan Sotelo, Christina Lee Ethridge, Kitra Cates, Robert Schmitz Department of Genetics | University of Georgia Transcription factors (TFs) are proteins that control the expression of specific genes. Investigating the interactions between TFs and their target DNA sequences benefits our understanding of how regulatory pathways are modulated. In humans, the improper regulation of transcription factors can lead to a number of diseases including diabetes and certain forms of cancer such as leukemia1. Traditionally, transcription factor binding motifs are determined using Chromatin Immunoprecipitation (ChIP)2. ChIP requires the use of specific antibodies to target DNA-TF interactions, and can be both costly and inefficient. Here, we utilize DNA Affinity Purification sequencing (DAP-seq), a novel in-vitro assay that is both economical and scalable3. This technique circumvents the traditional limitations of ChIP-seq and provides a feasible alternative to studying DNA-TF interactions. Abstract Motif Analysis of DAP-seq Acknowledgements References Next Steps We would like to thank the National Science Foundation and start-up funds from the University of Georgia for supporting this study, along with Michael Gonzales (UGA Outreach Program Coordinator). 1. Villard, Jean. "Transcription regulation and human diseases." Swiss medical weekly 134 (2004): 571-579. 2. Schmidt, D., Wilson, M. D., Spyrou, C., Brown, G. D., Hadfield, J., & Odom, D. T. (2009). ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods (San Diego, Calif.), 48(3), 240–248. 3. O’Malley, R., Huang, S., Song, L., Lewsey, M., Bartlett, A., Nery, J., . . . Ecker, J. (2016). Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell, 165(5), 1280-1292. 4. Mönke G, Seifert M, Keilwagen J, et al. Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Research. 2012;40(17):8240- 8254. Methods • Scale DAP-seq up to a 96-well plate format • Analyze all TFs in the maize and soybean genomes • Determine all currently unknown TF binding motifs in the cistrome of Arabidopsis thaliana, including those of TFs that operate during heat shock, drought, and other abiotic stressors • Map out gene regulatory networks controlled by TFs. • Alter and manipulate properties in gene regulatory networks using techniques such as CRISPR Figure 1: A plasmid containing the transcription factor sequence and the Halo affinity tag is transcribed and translated in vitro. This produces a fusion protein consisting of the transcription factor and the Halo tag. DAP-seq Results for ABI3 Figure 2: Interaction between genomic DNA, a TF-Halo fusion protein, and a magnetic bead. The transcription factor binds sonicated genomic DNA. The Halo tag is then covalently bound to the magnetic bead. This interaction allows for the pull down of TF-bound DNA for subsequent sequencing. Figure 5 (Above): A magnified image of an ABI3 transcription factor peak from Figure 3C. In this region, we identified the binding motif for ABI3 to be CATGCA (outlined in yellow). Figure 4 (Below): Sequence logo of the ABI3 transcription factor binding motif representing a genome-wide assessment of ABI3 binding domains from ChIP-seq4. 3A Figure 3: Visualization of DAP-seq results for Zea mays transcription factor ABI3. The orange peaks with black tips represent regions of the genome where this transcription factor bound. In accordance with our expectation, these peaks align with the promoter regions of various Z. mays genes. 3B 3C