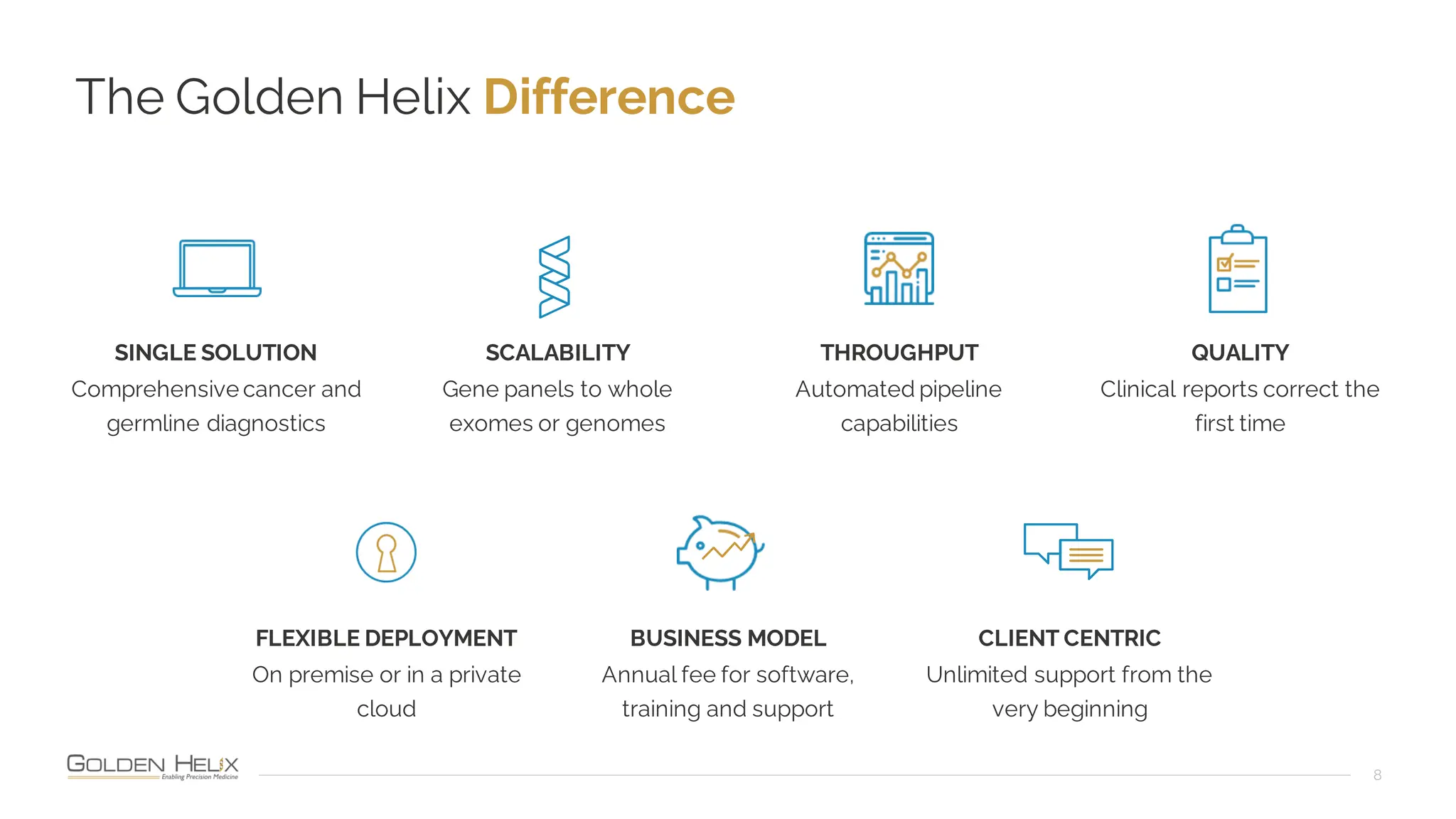
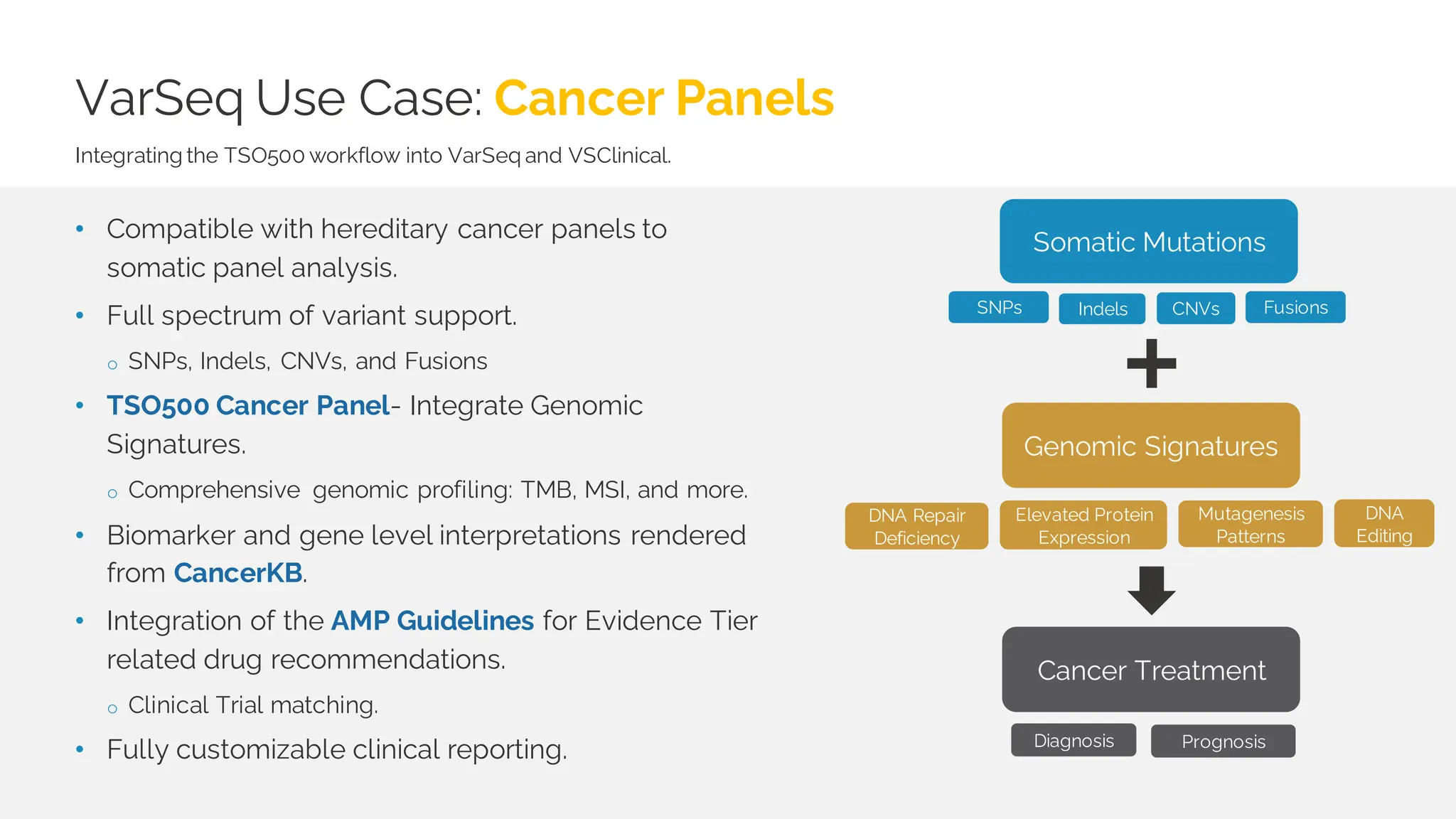
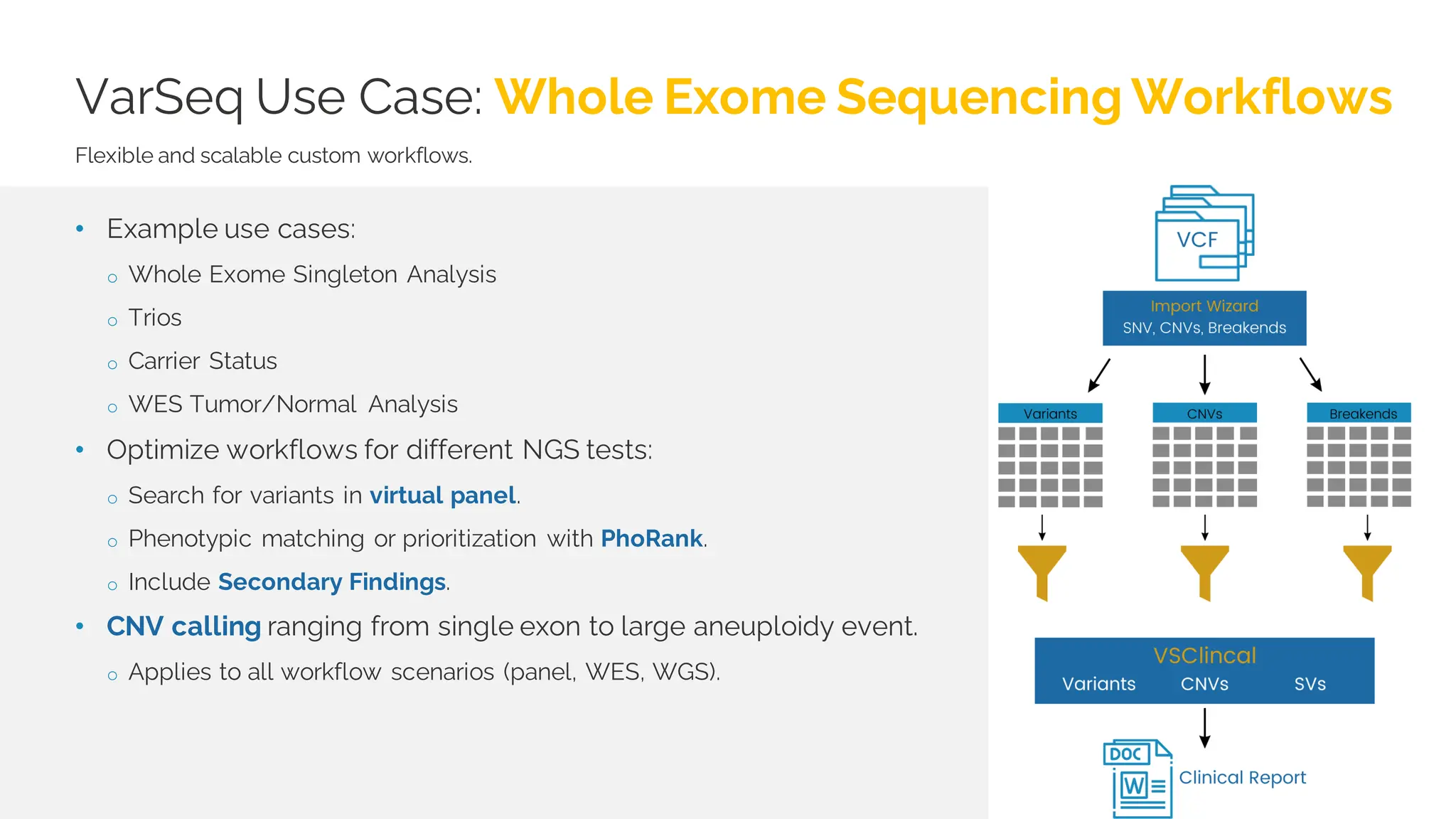
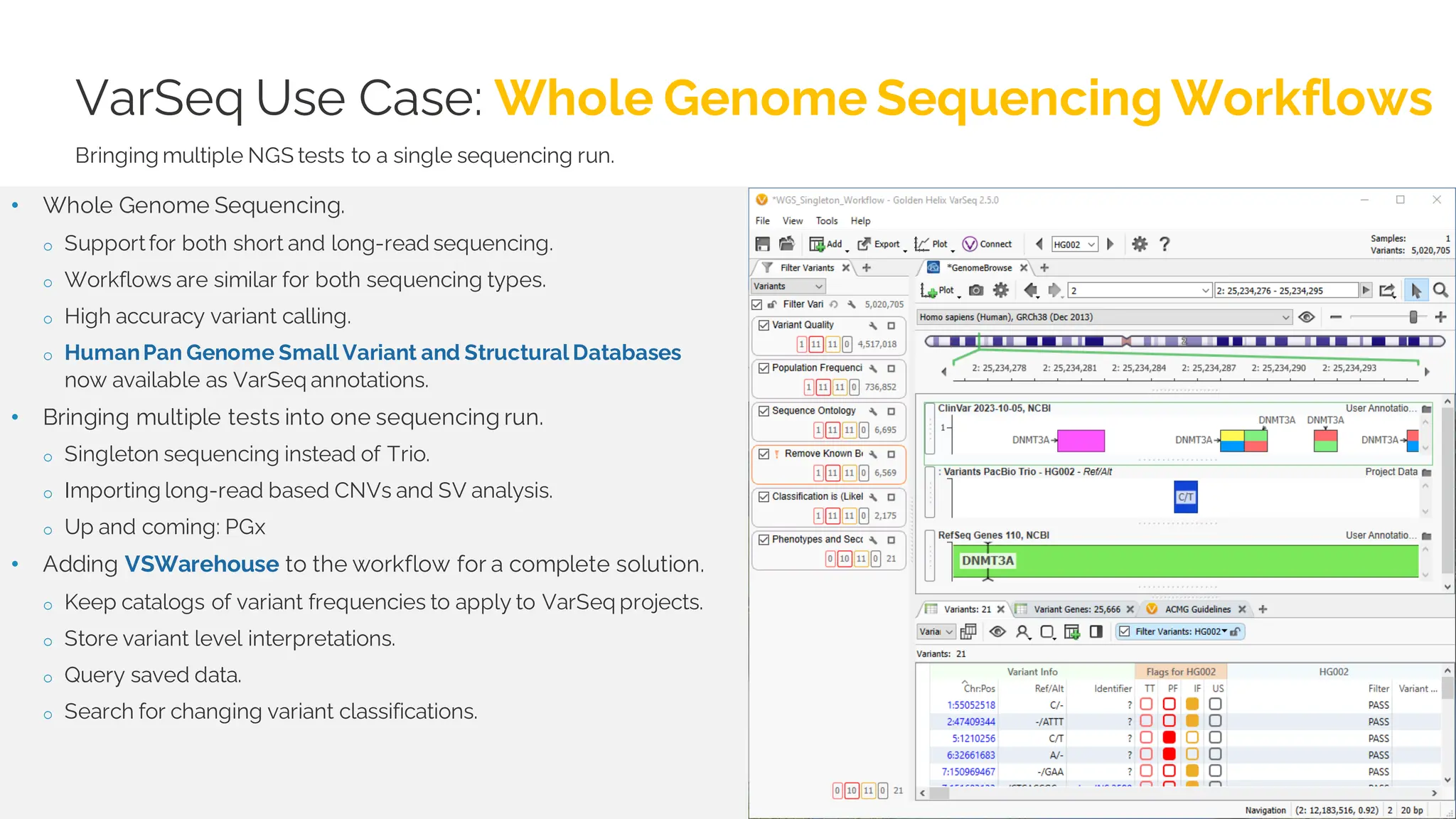
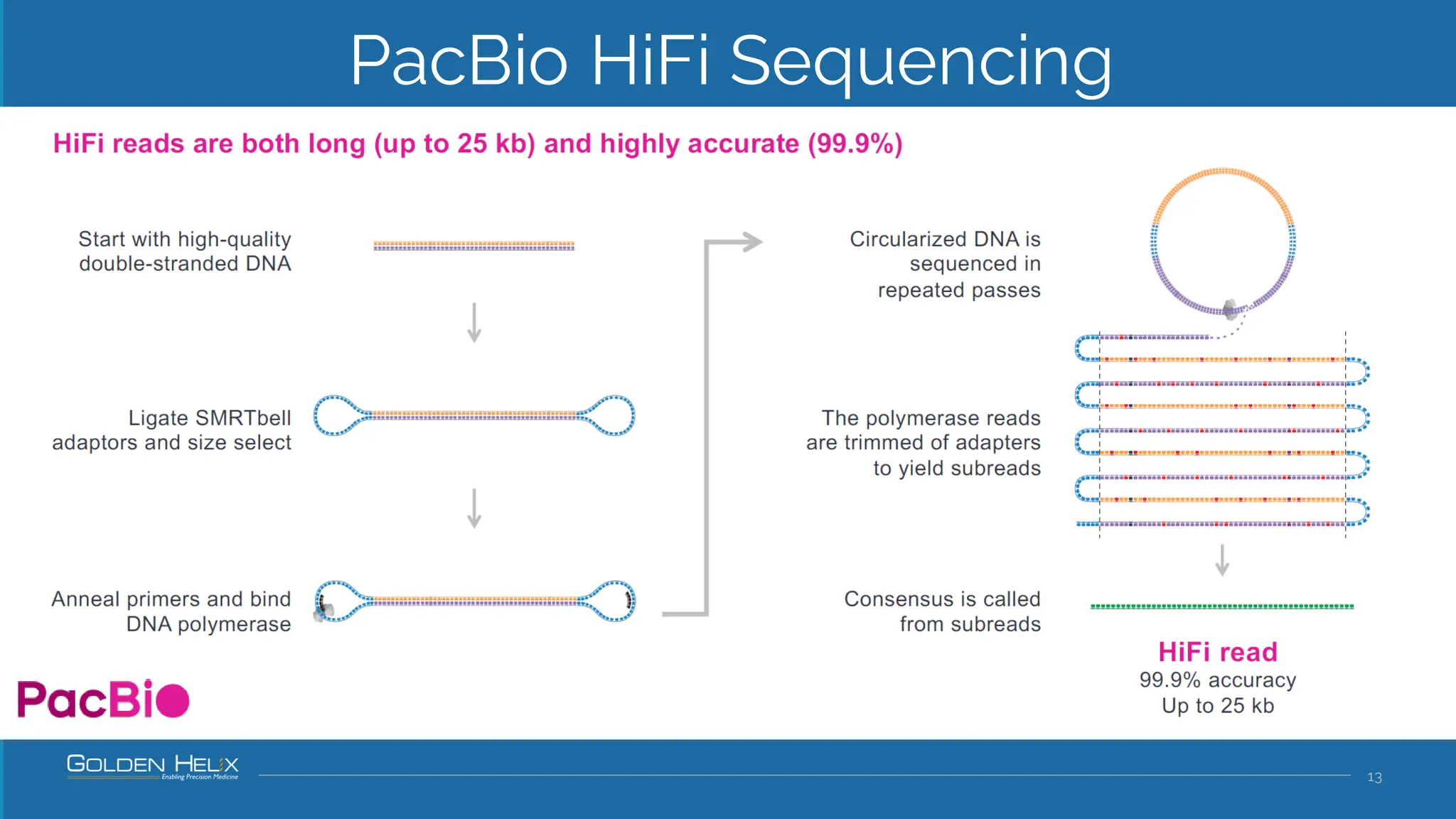
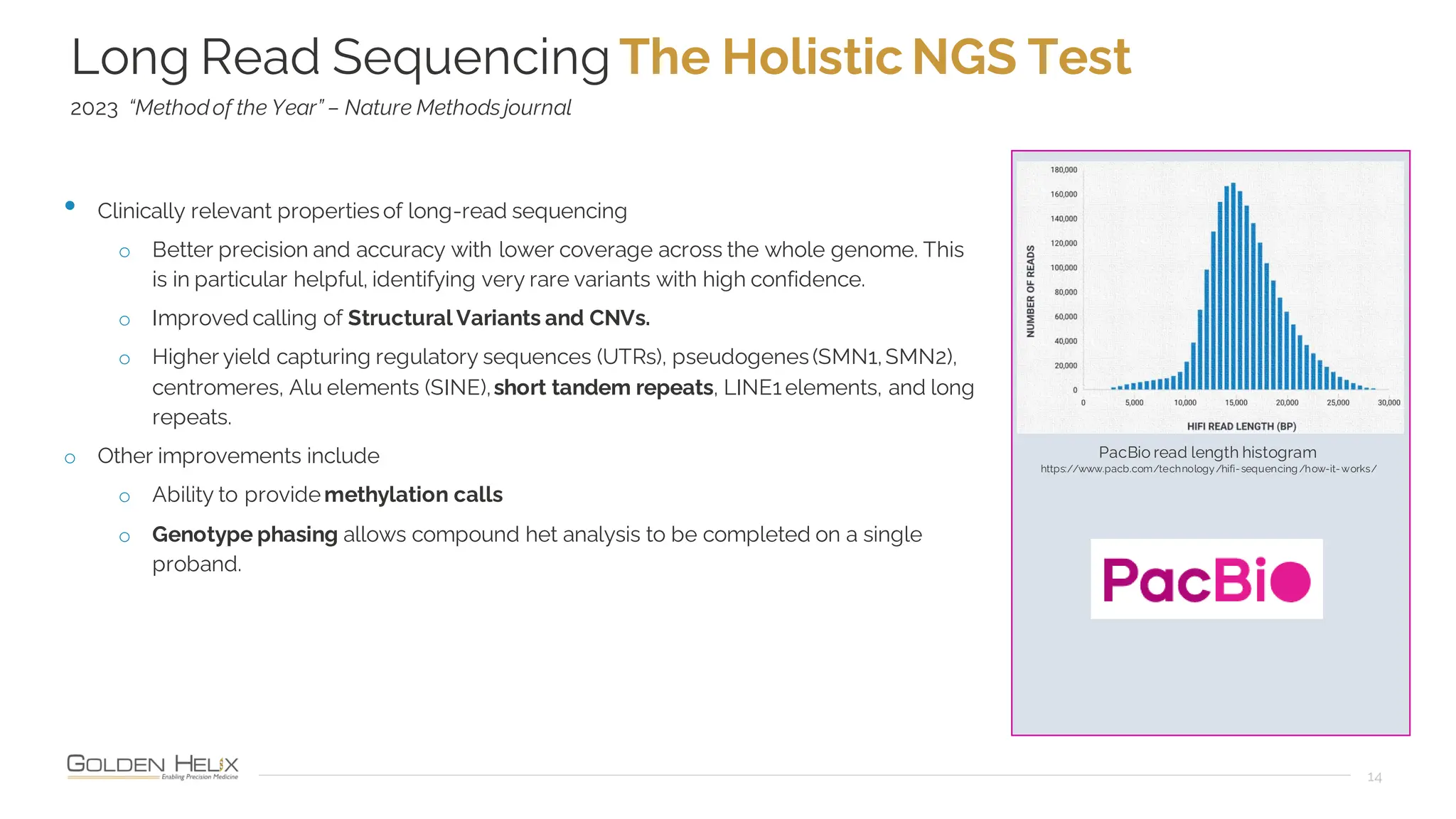
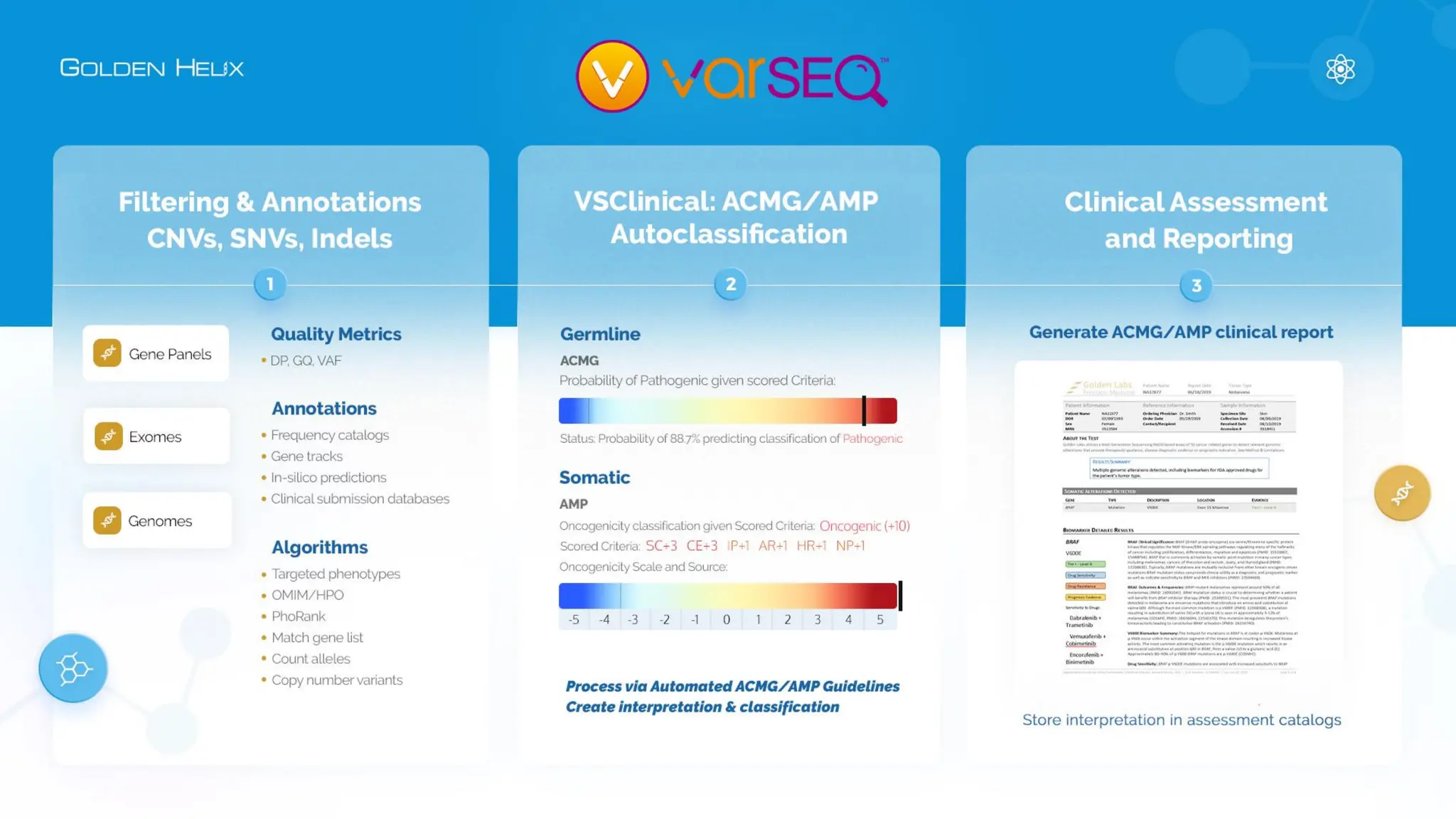
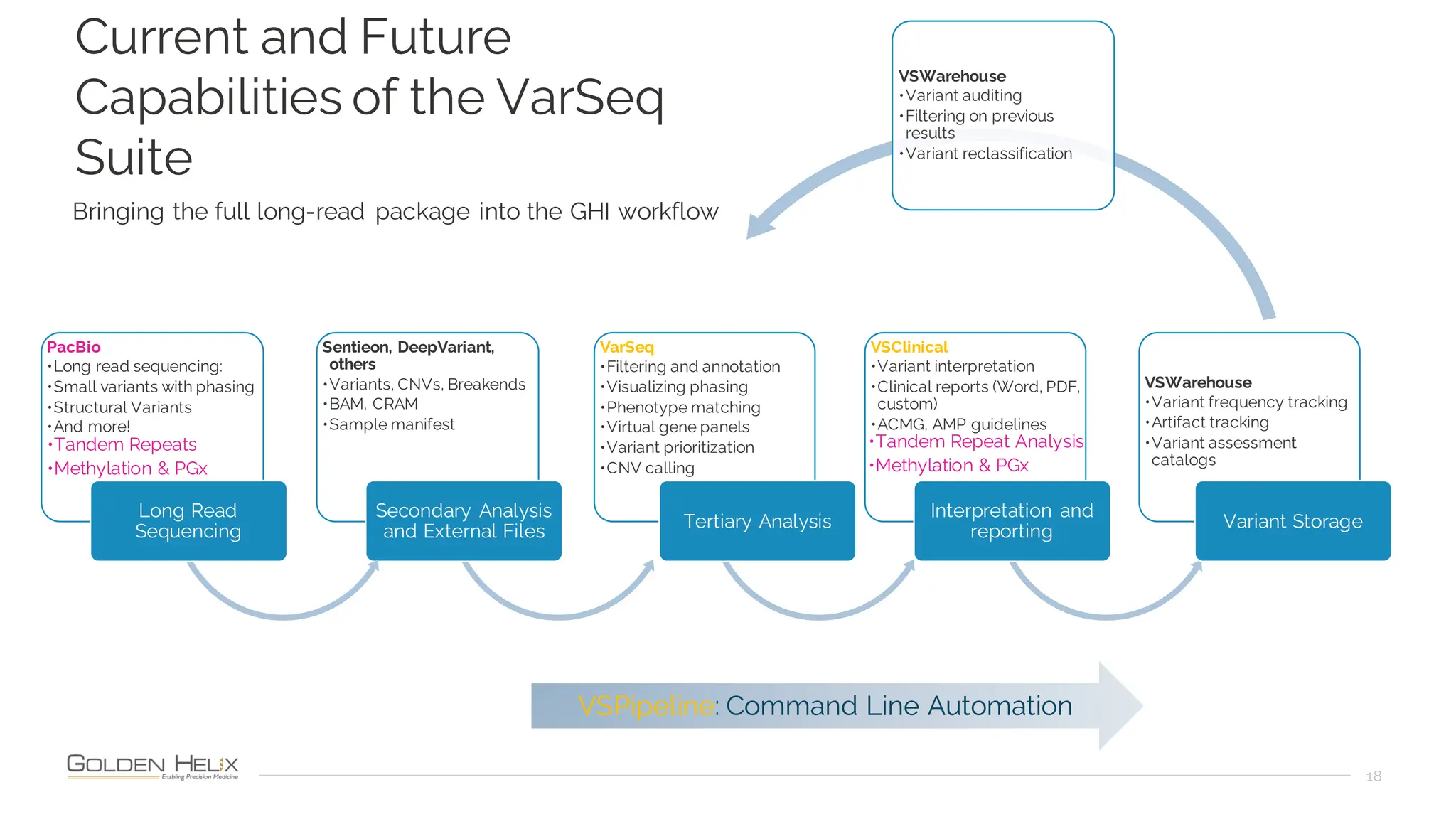
The document details a presentation by Dr. Jennifer Dankoff on utilizing VarSeq, a bioinformatics platform by Golden Helix, for short and long-read next-generation sequencing (NGS) data, particularly in cancer genetics. It highlights the integration of various workflows such as cancer panels, whole exome sequencing, and whole genome sequencing, emphasizing features like variant calling, customizable reporting, and compliance with ACMG and AMP guidelines. The presentation also covers the benefits of long-read sequencing technologies and their role in enhancing genomic profiling and analysis capabilities.