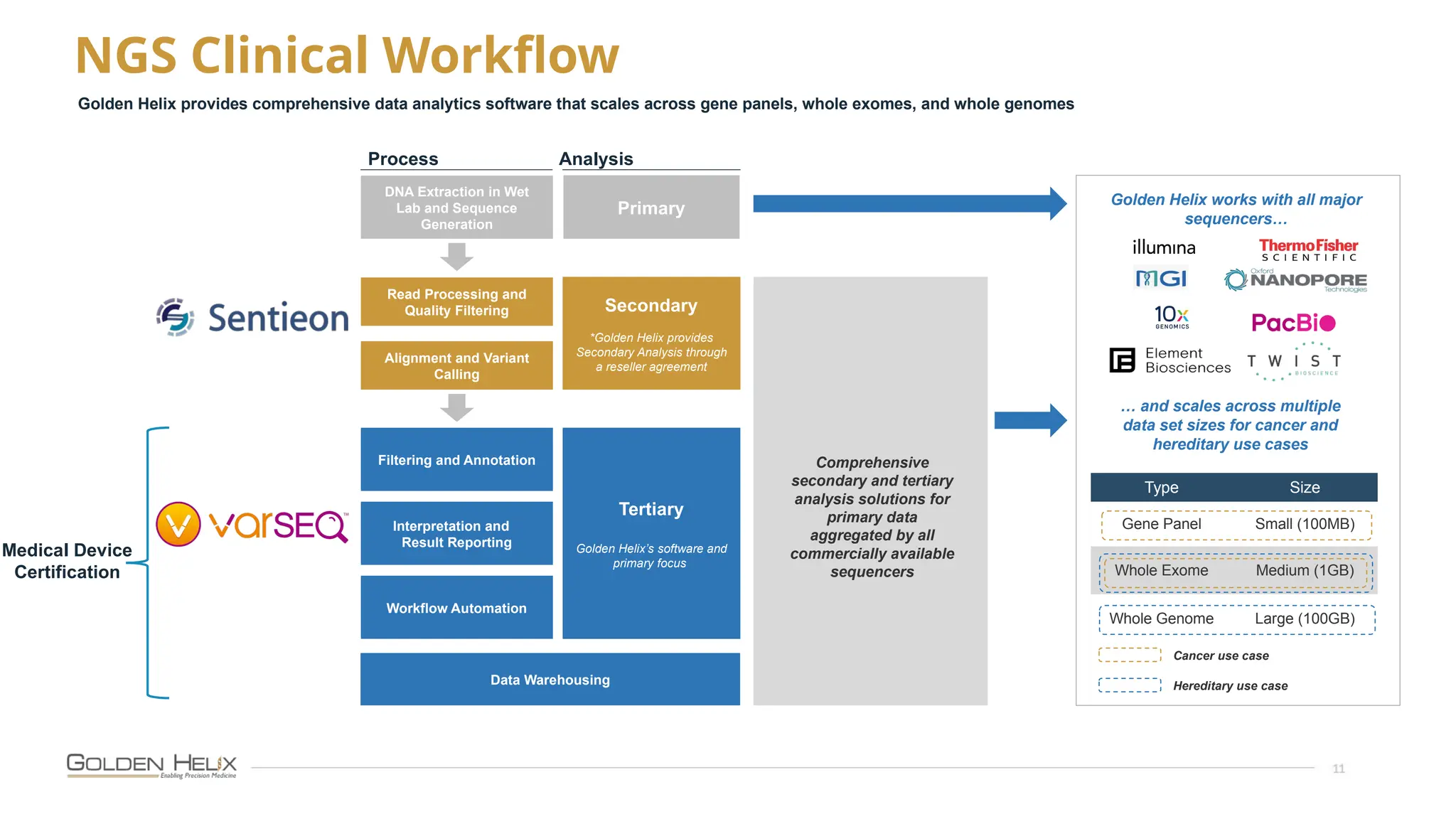
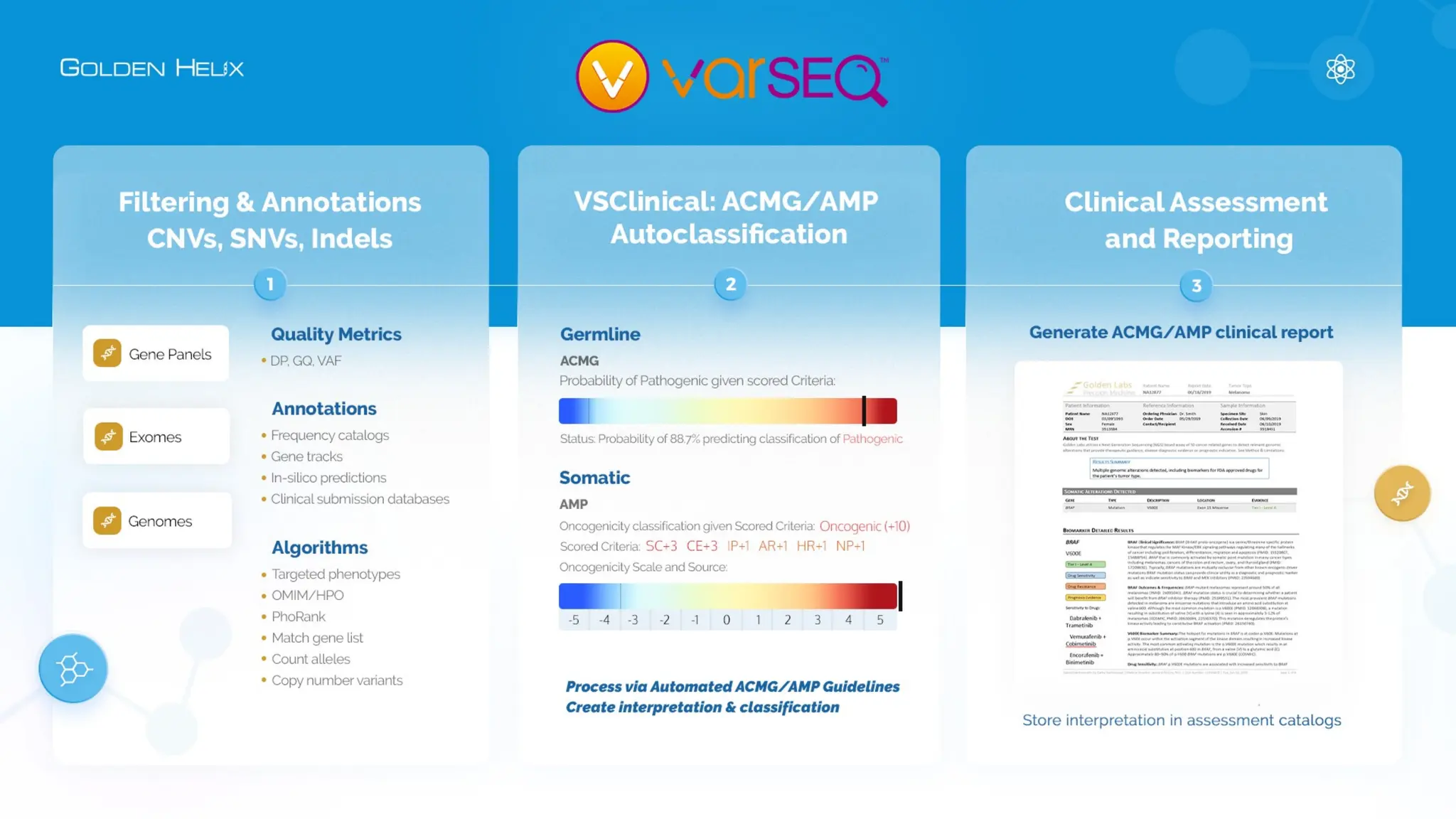
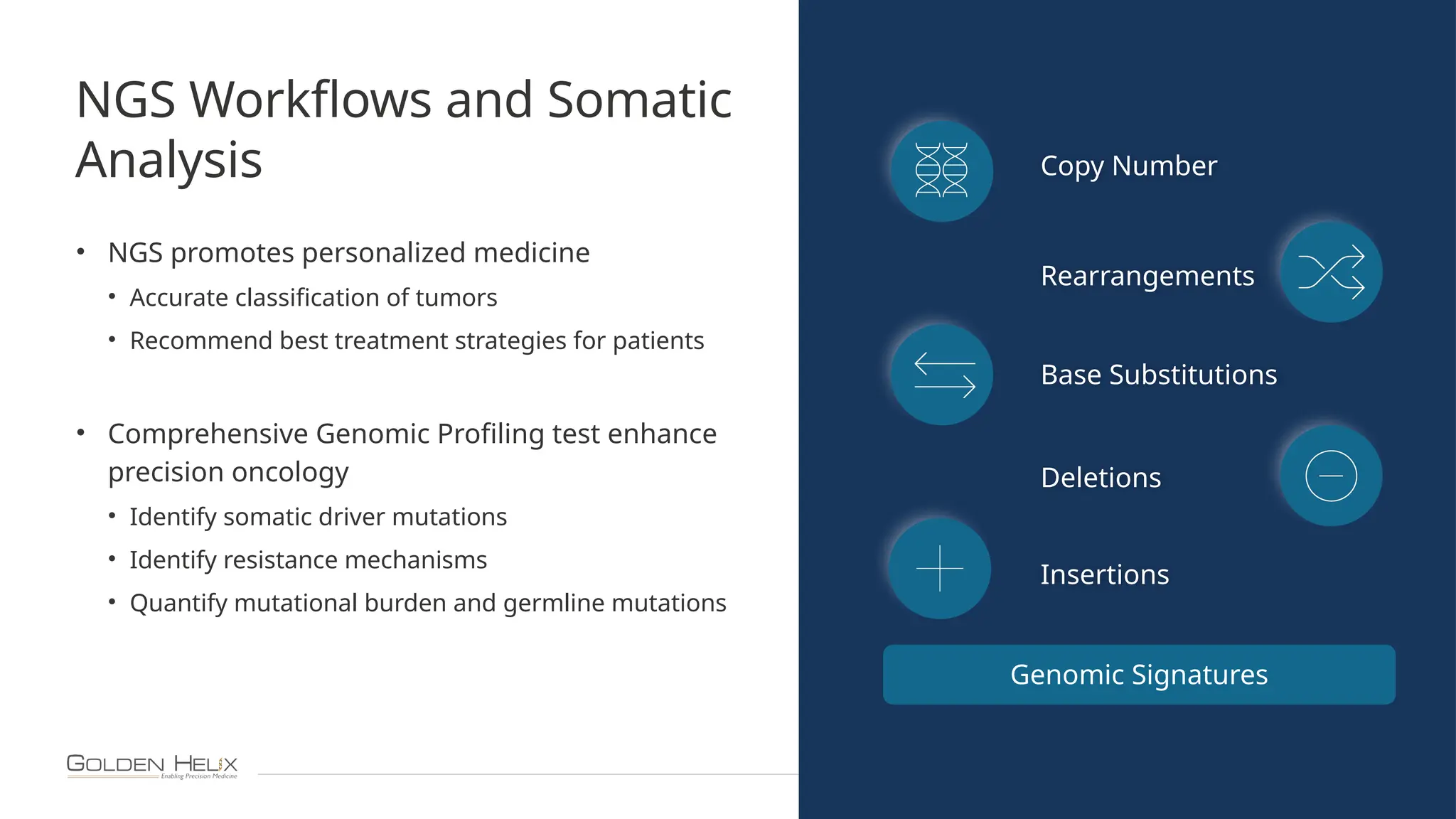
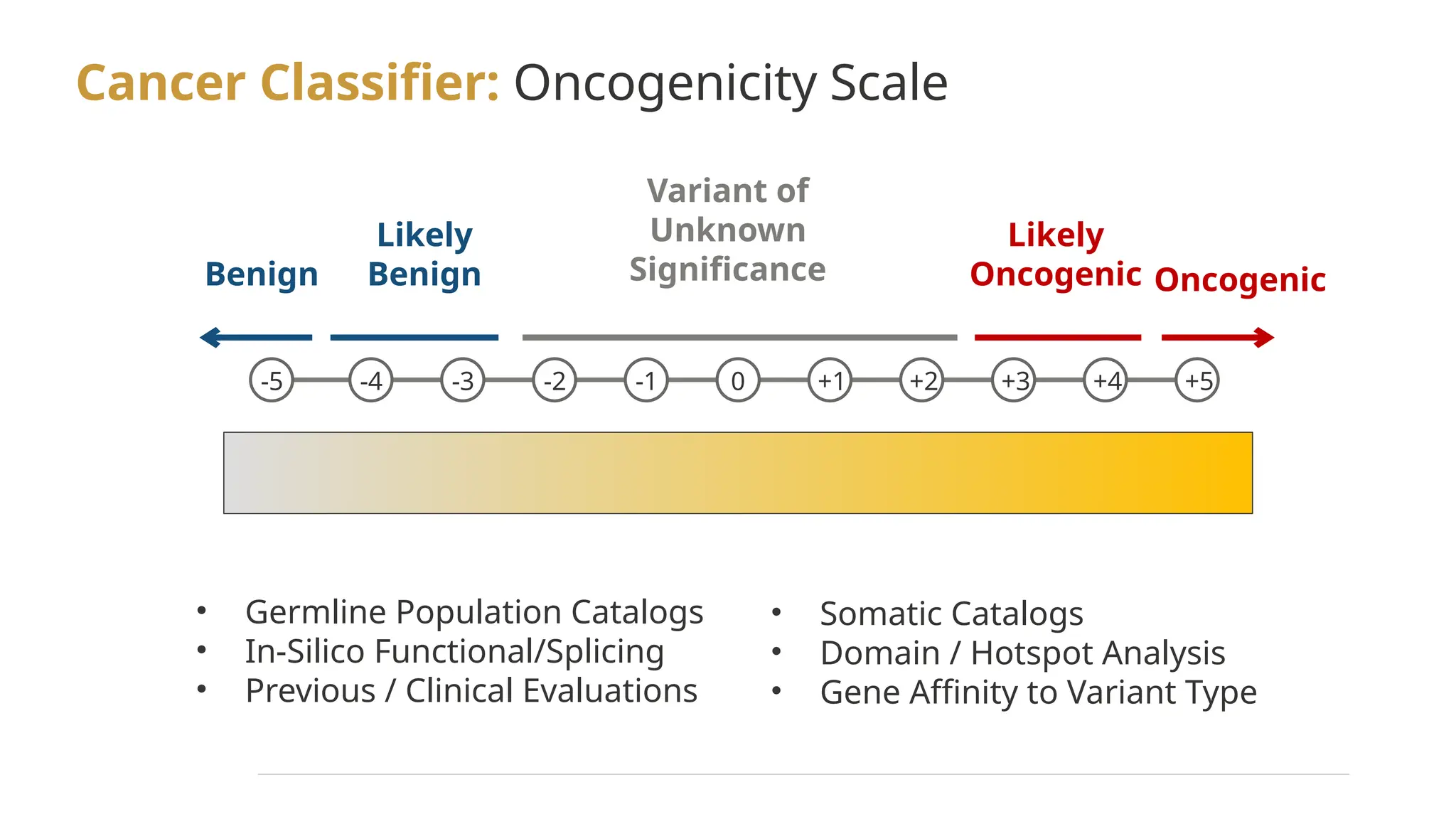
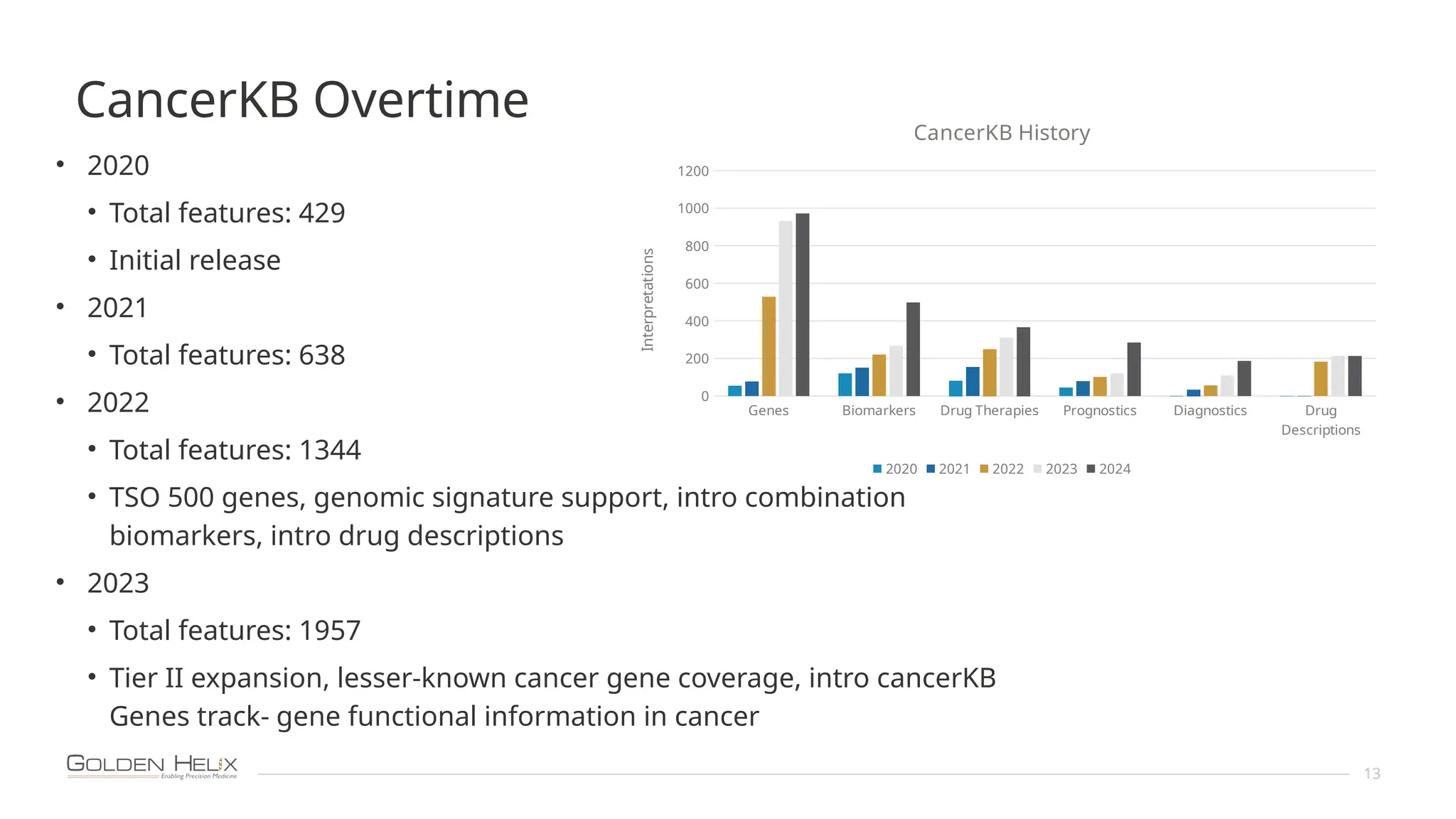
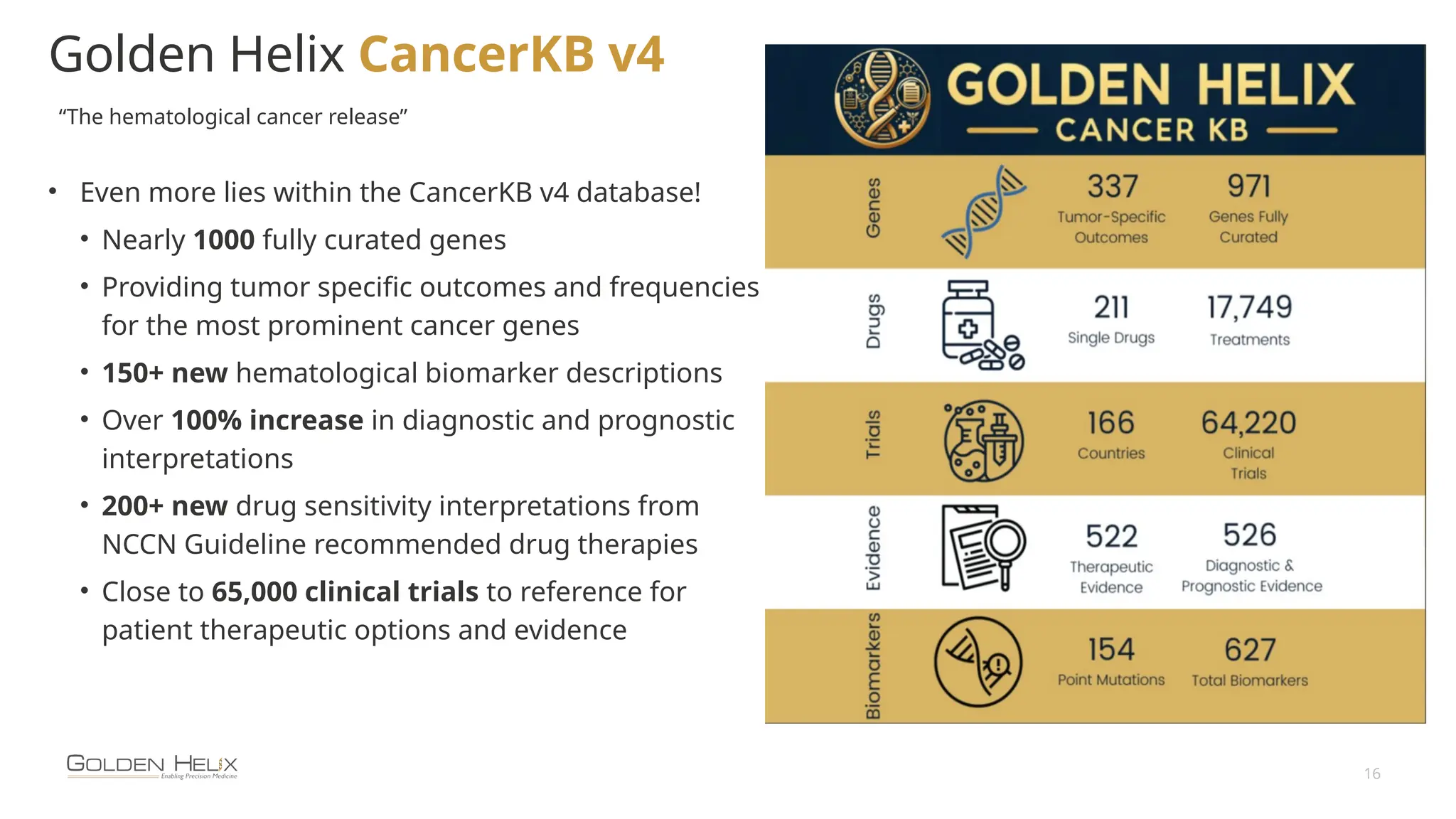
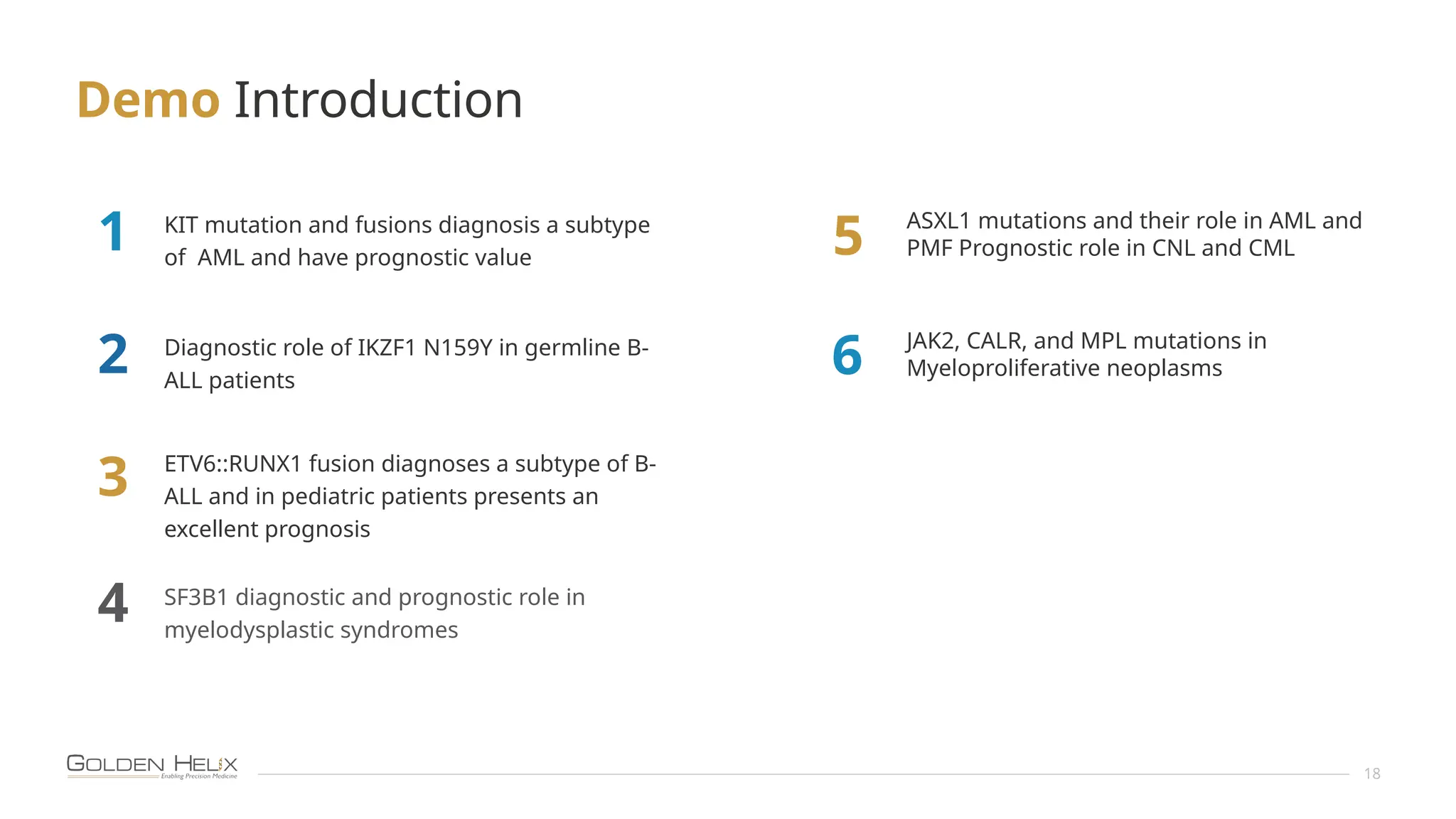
The document details the features and advancements of Golden Helix's CancerKB 4.0 and somatic analysis within its VSClinical software, highlighting its compliance with European regulations and support for clinical genomic analysis. It emphasizes the database's role in cancer variant interpretation, particularly for hematological malignancies, and mentions various NIH grants supporting the research. Additionally, it promotes the company's comprehensive eBook library and automated workflows aimed at improving genomic proficiency within clinical settings.