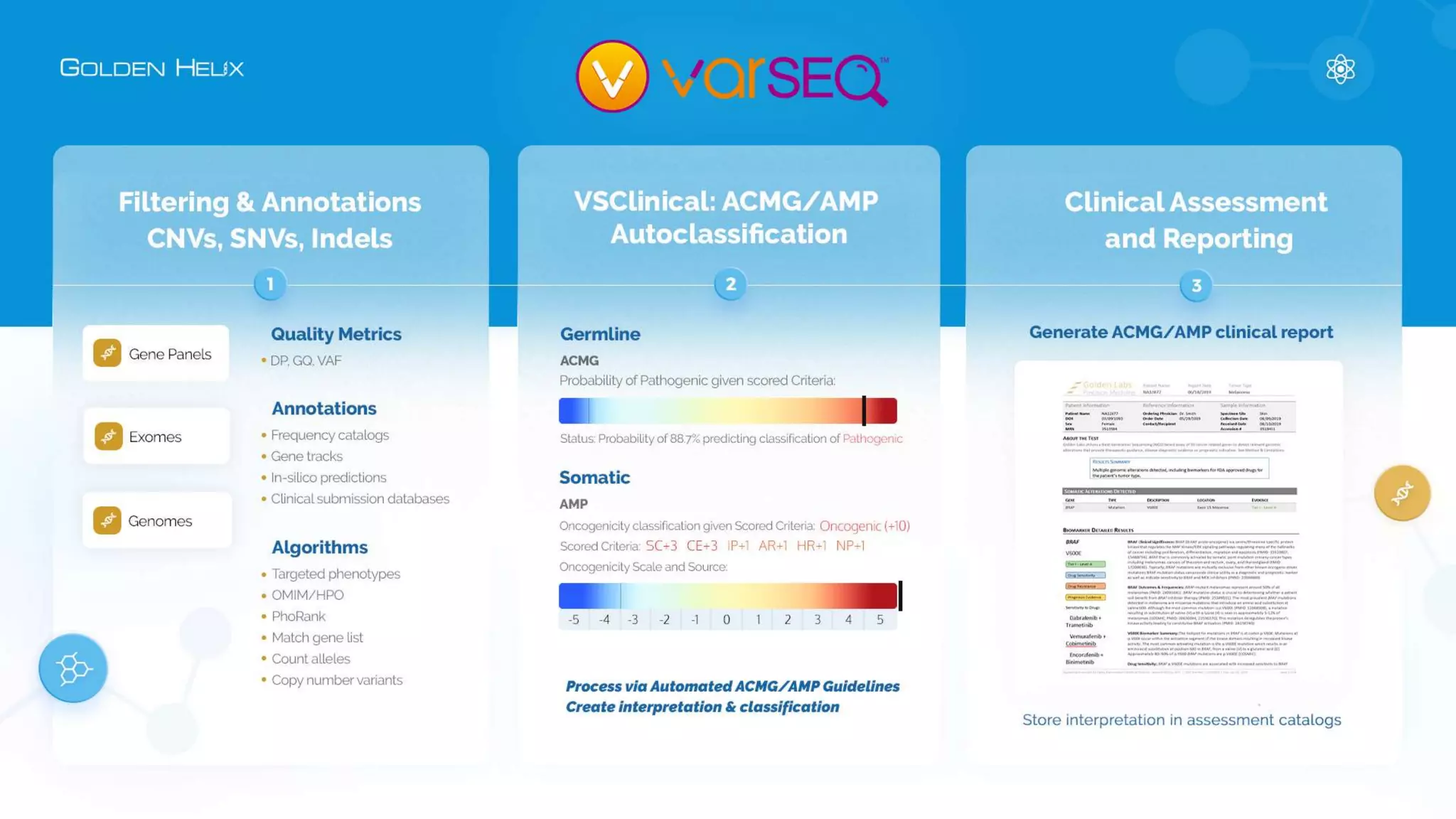
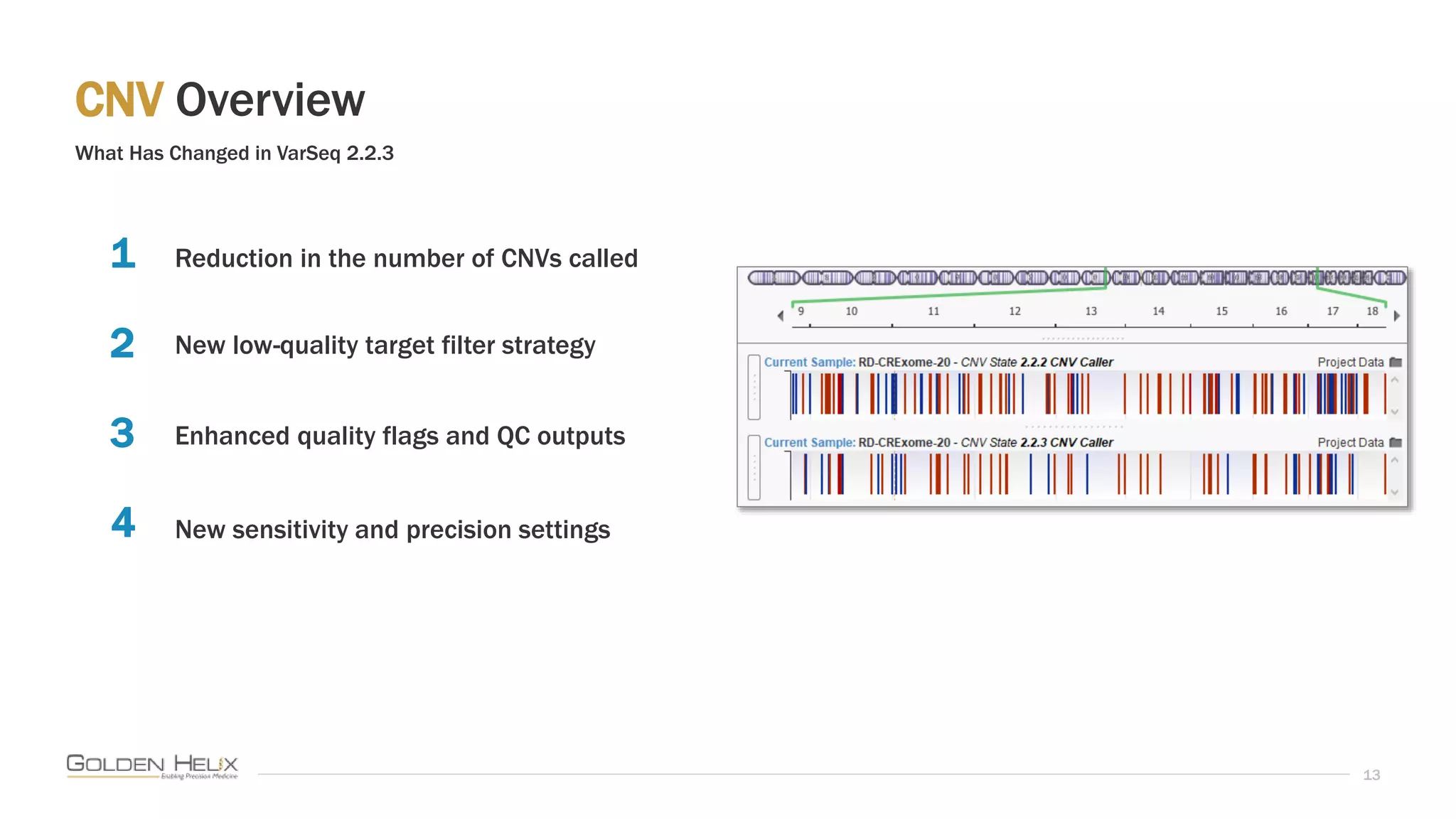
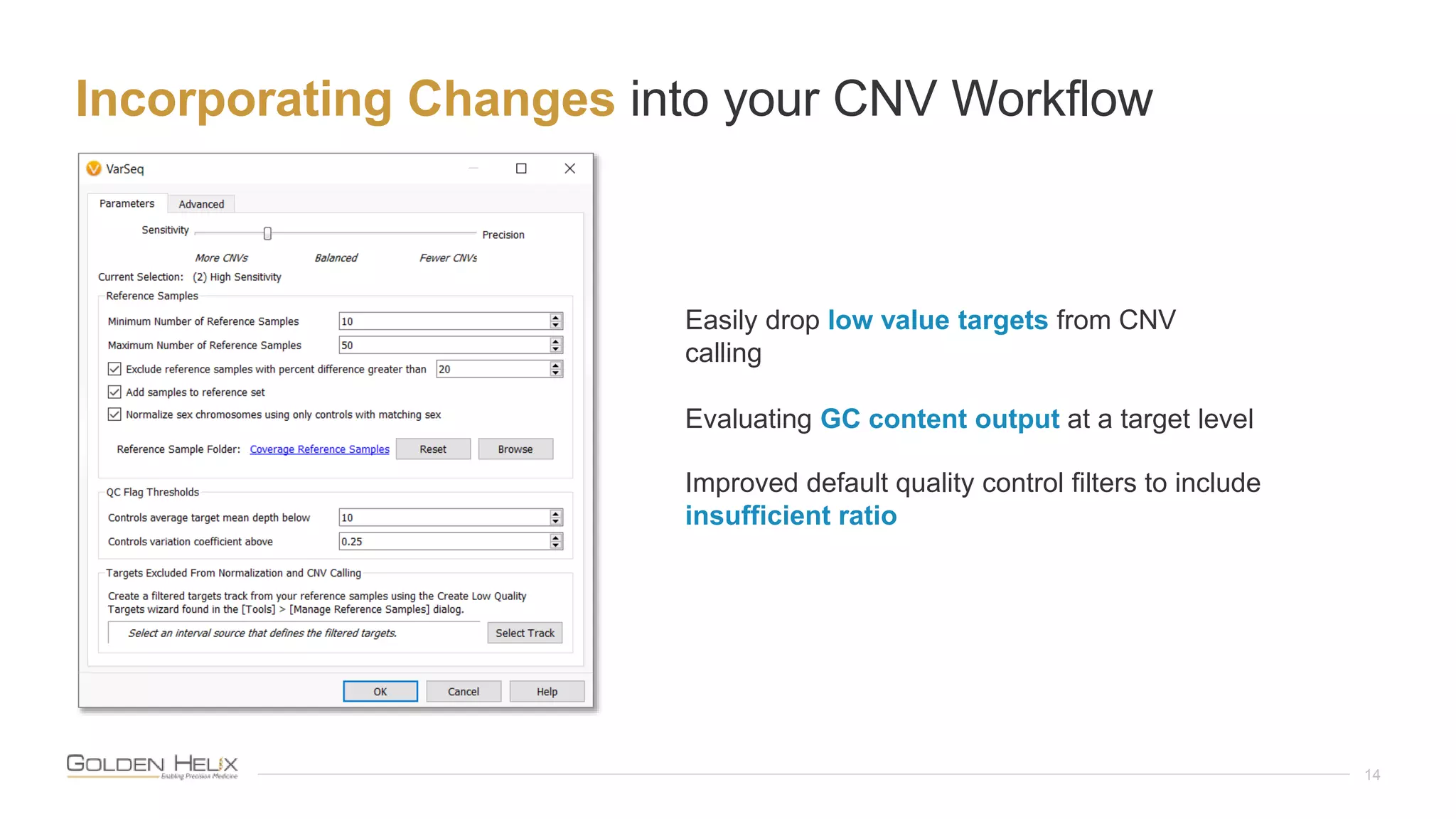
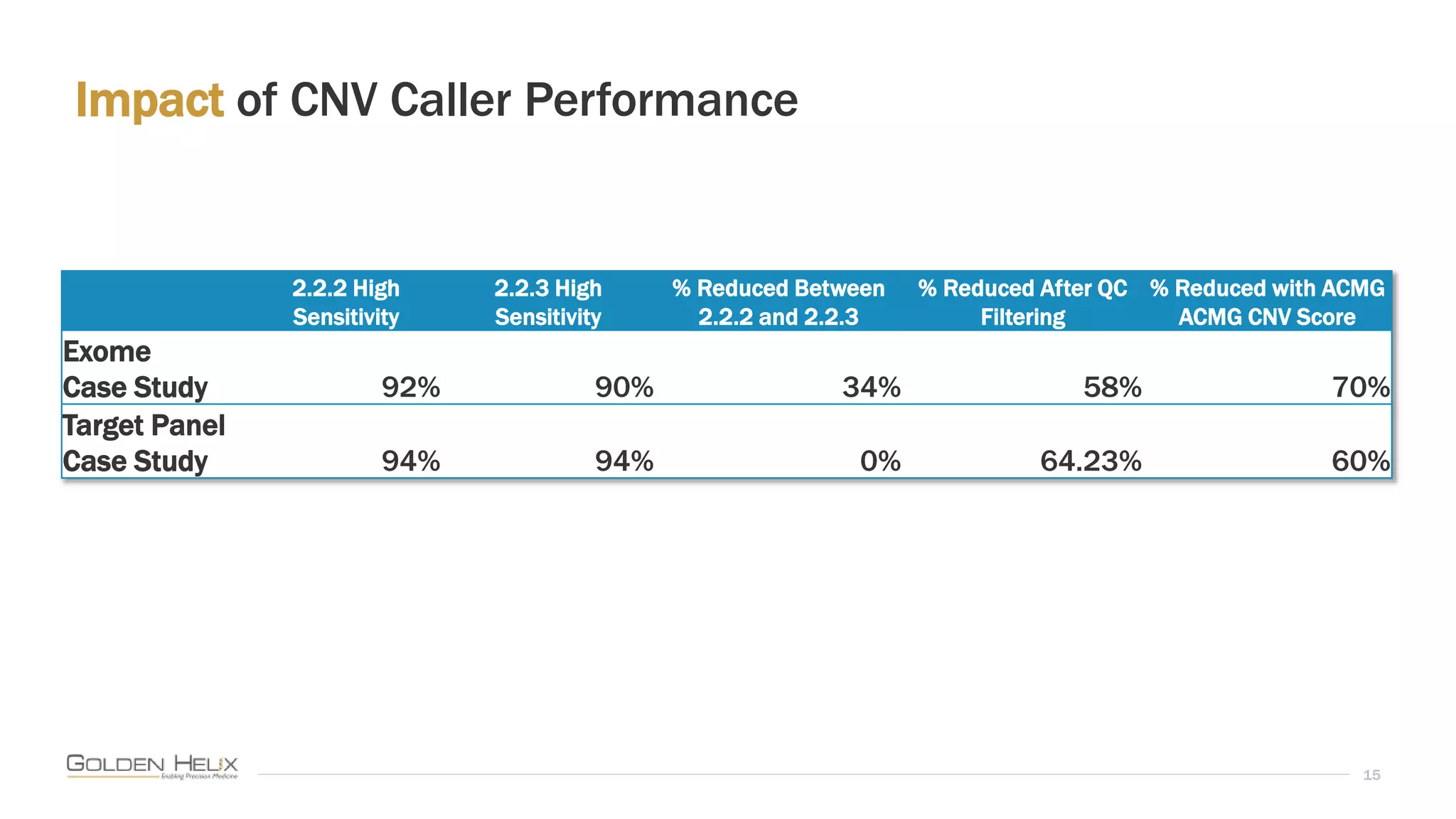
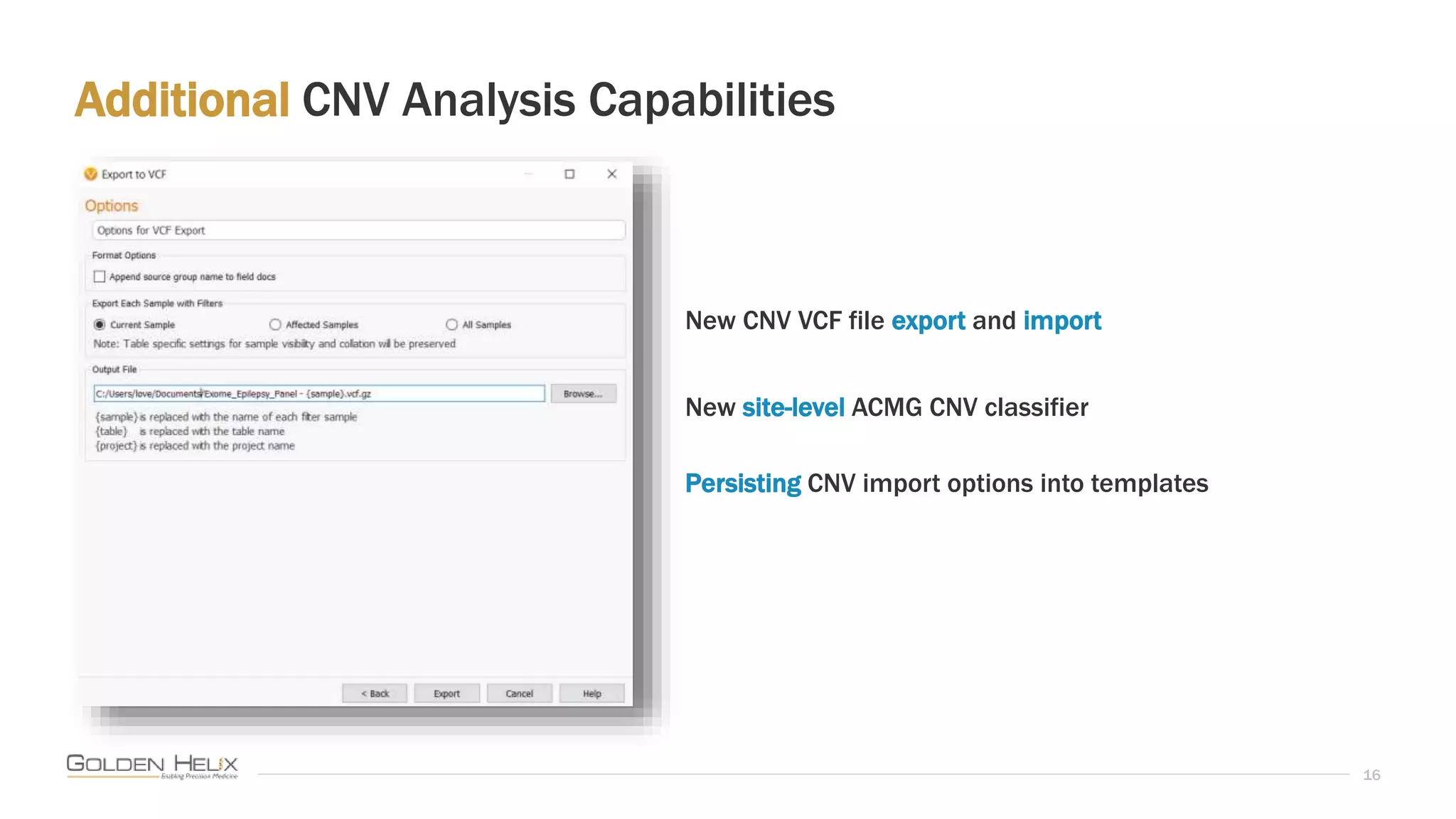
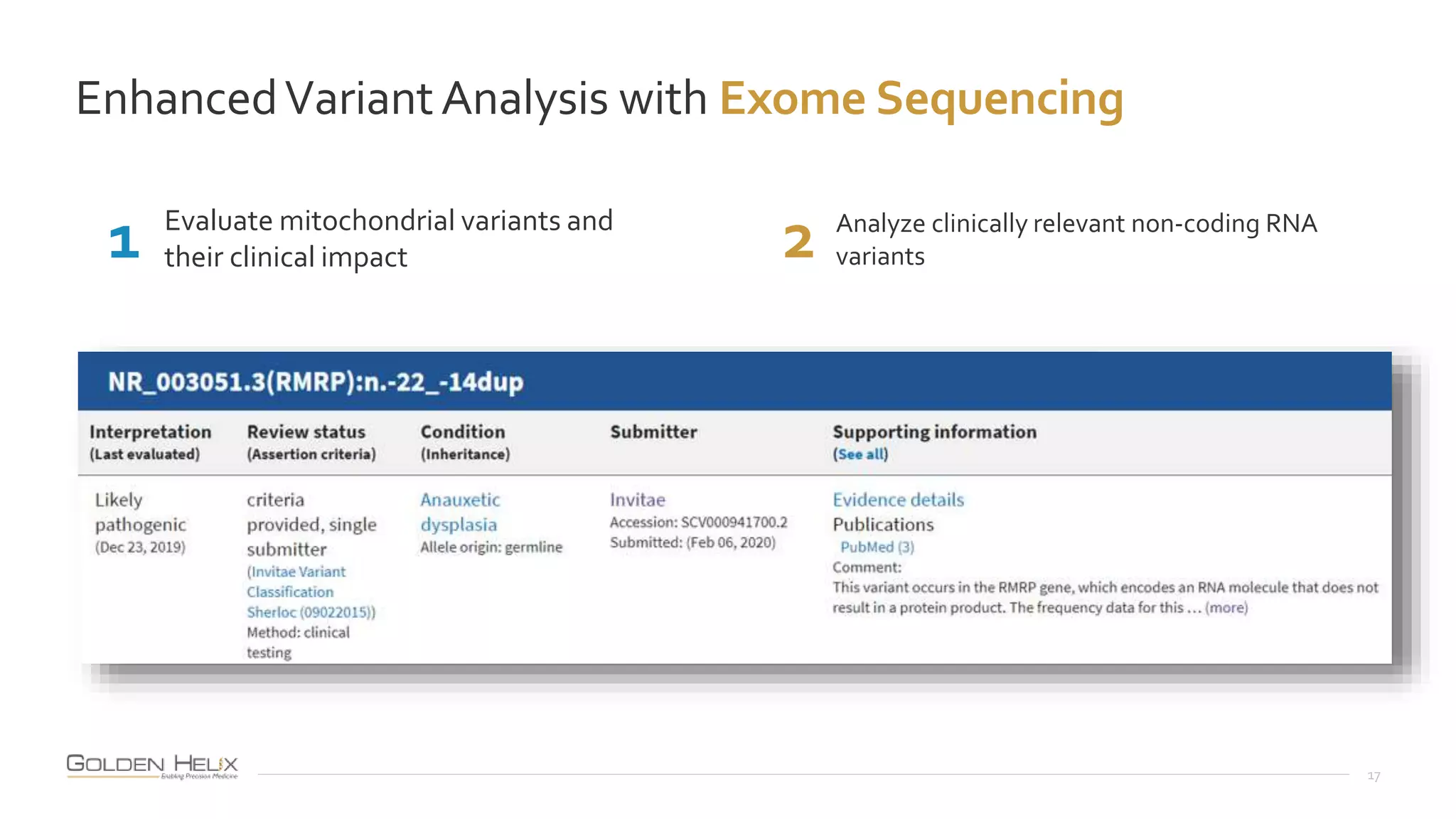
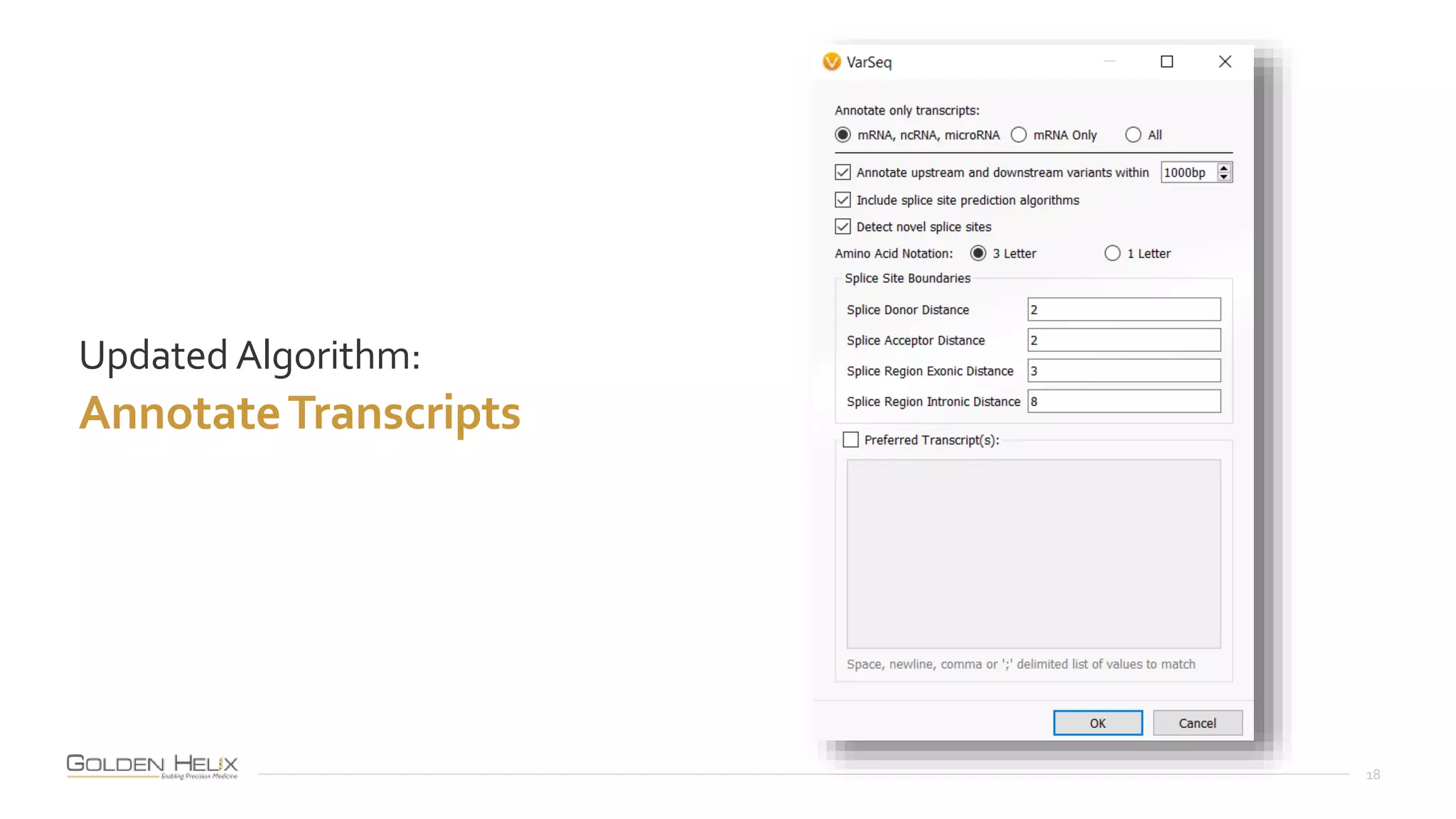
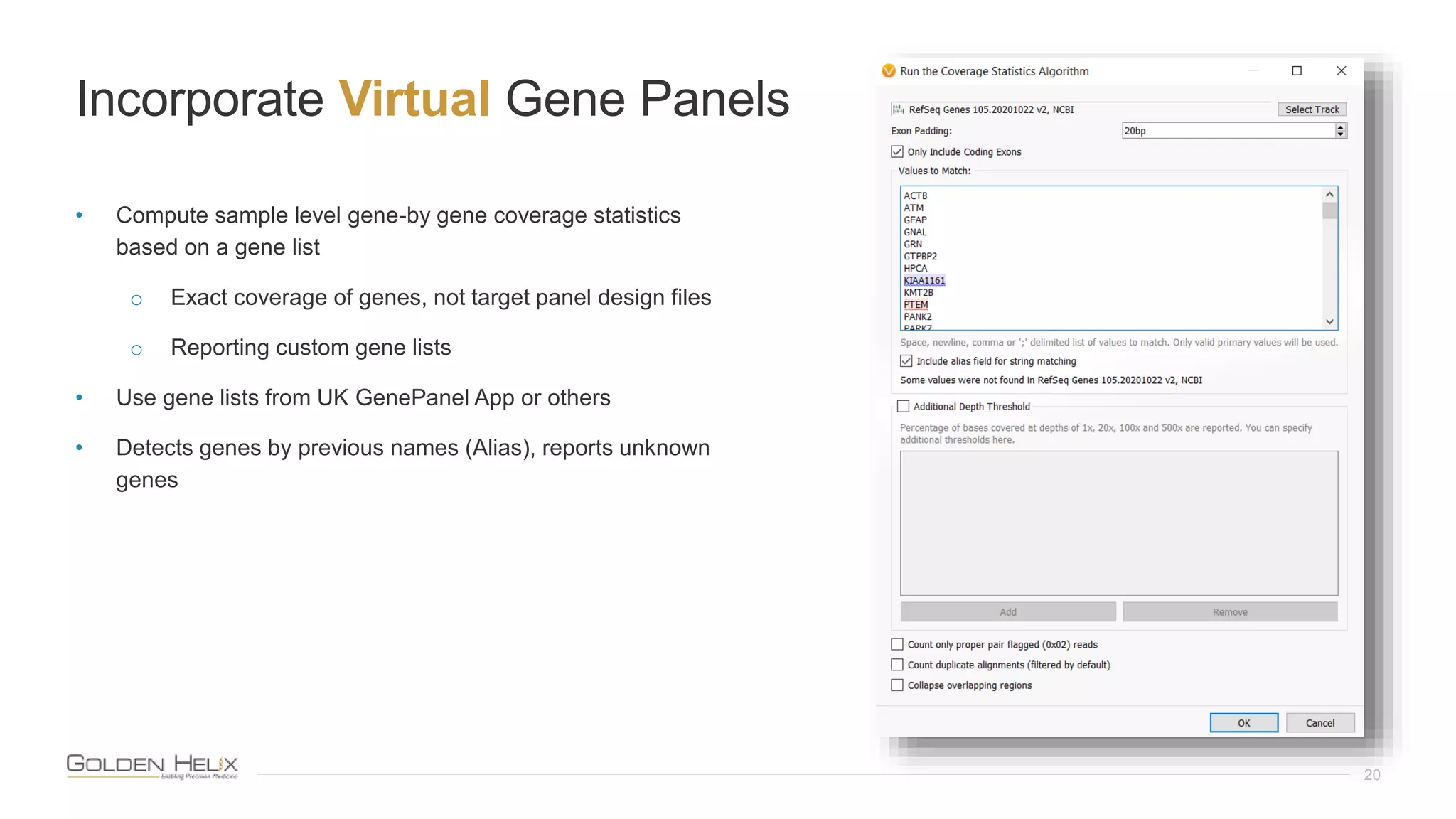
This document summarizes a presentation given by Gabe Rudy, VP of Product & Engineering at Golden Helix, on updated strategies and expanded capabilities for exome analysis with VS-CNV and VSClinical. The presentation demonstrates new features for enhanced variant analysis, including analyzing splice site, non-coding RNA, and mitochondrial variants. It also reviews improvements to Golden Helix's exome CNV calling in VarSeq, including a new low-quality target filter strategy that reduces the number of CNVs called while maintaining sensitivity and precision. Additional capabilities discussed include generating synchronized clinical reports and incorporating virtual gene panels.