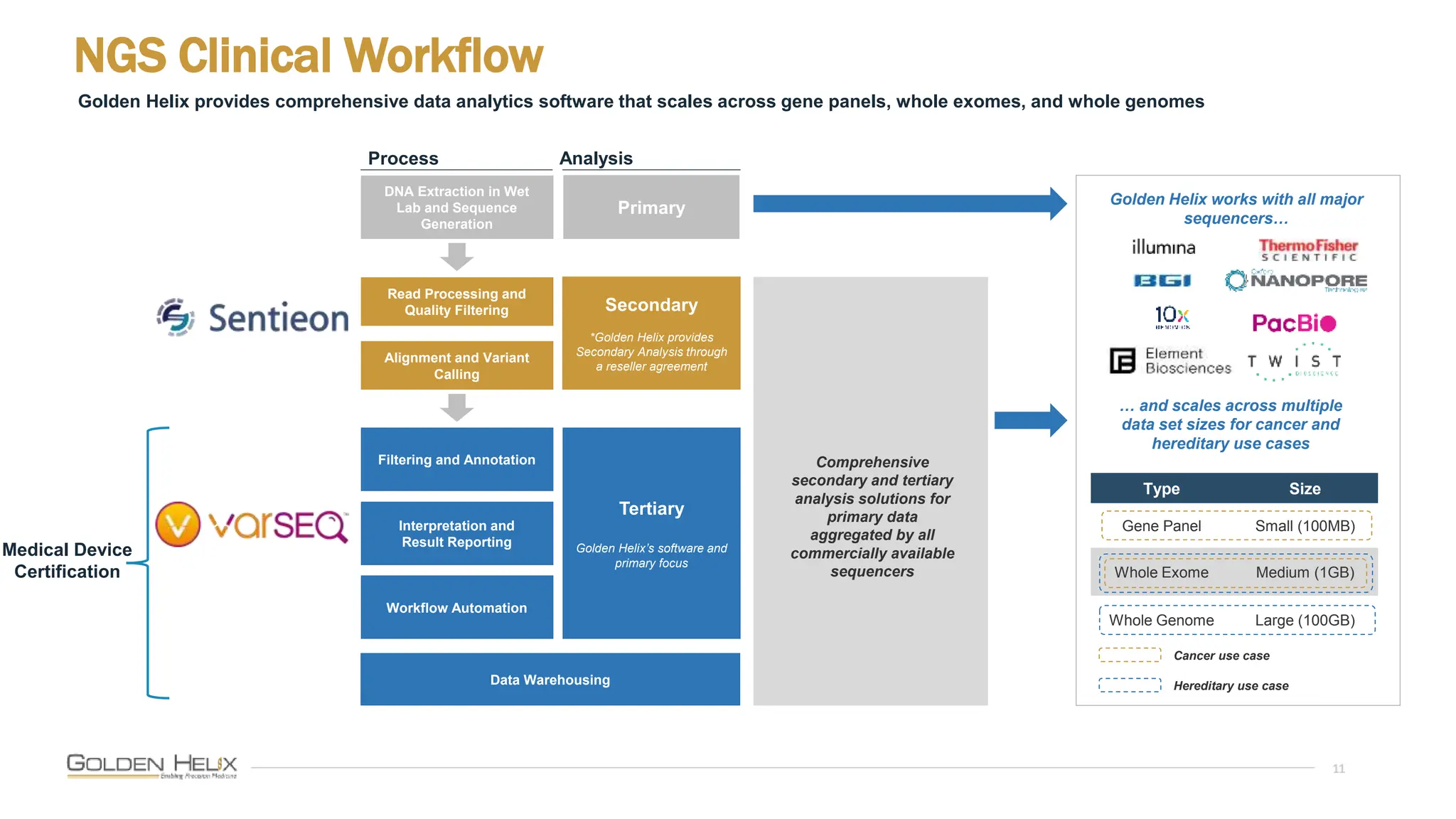
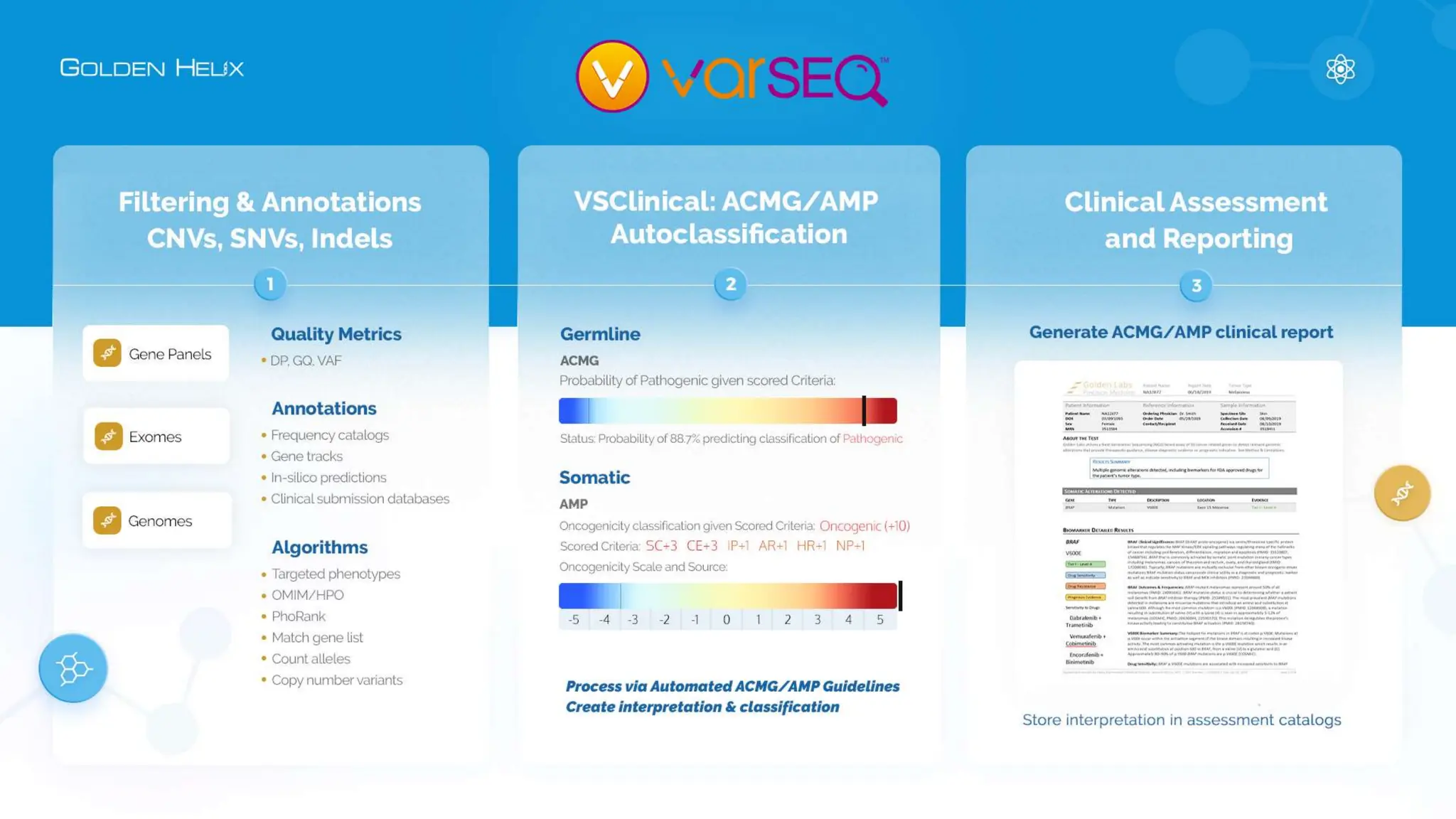
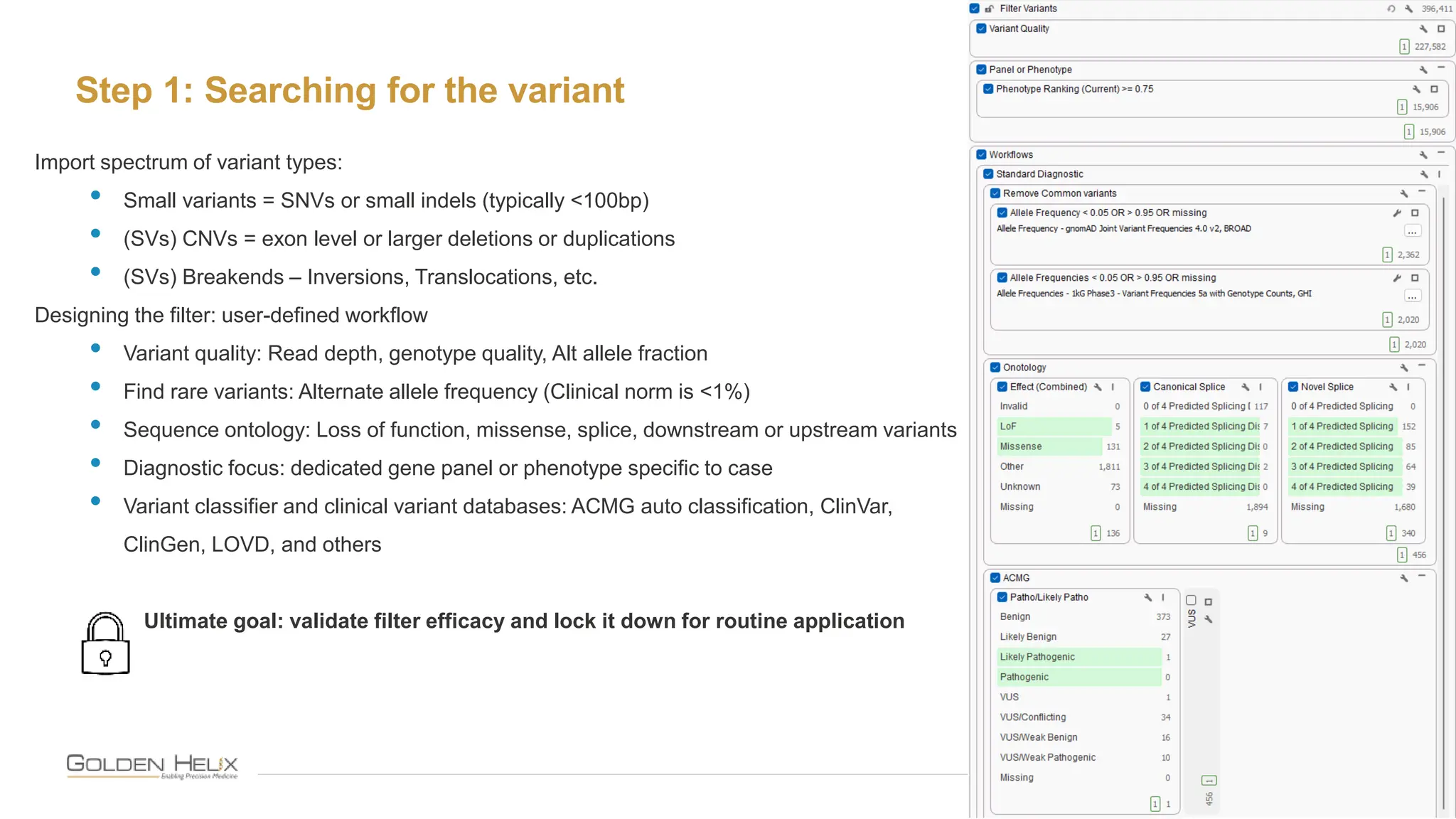
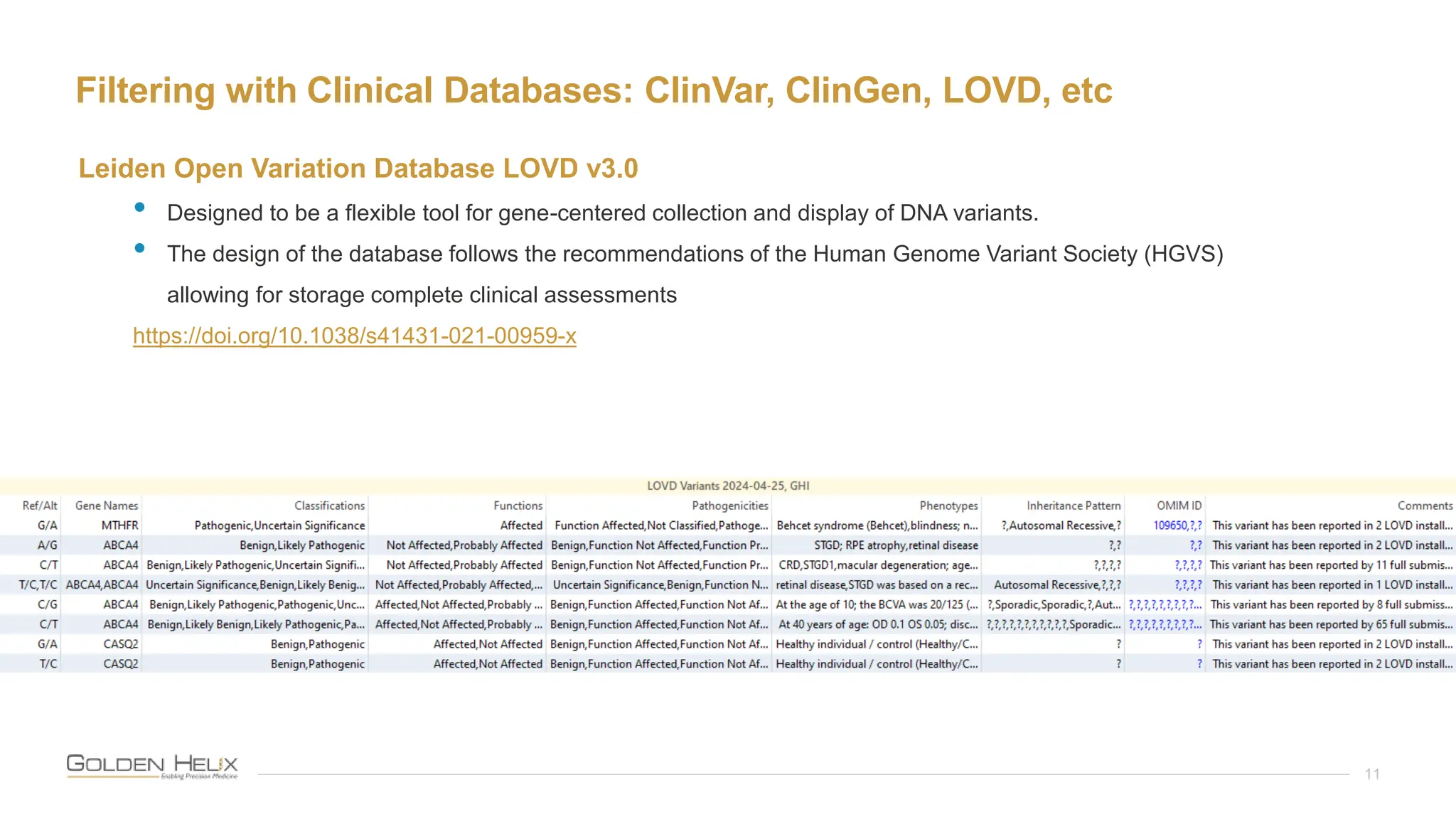
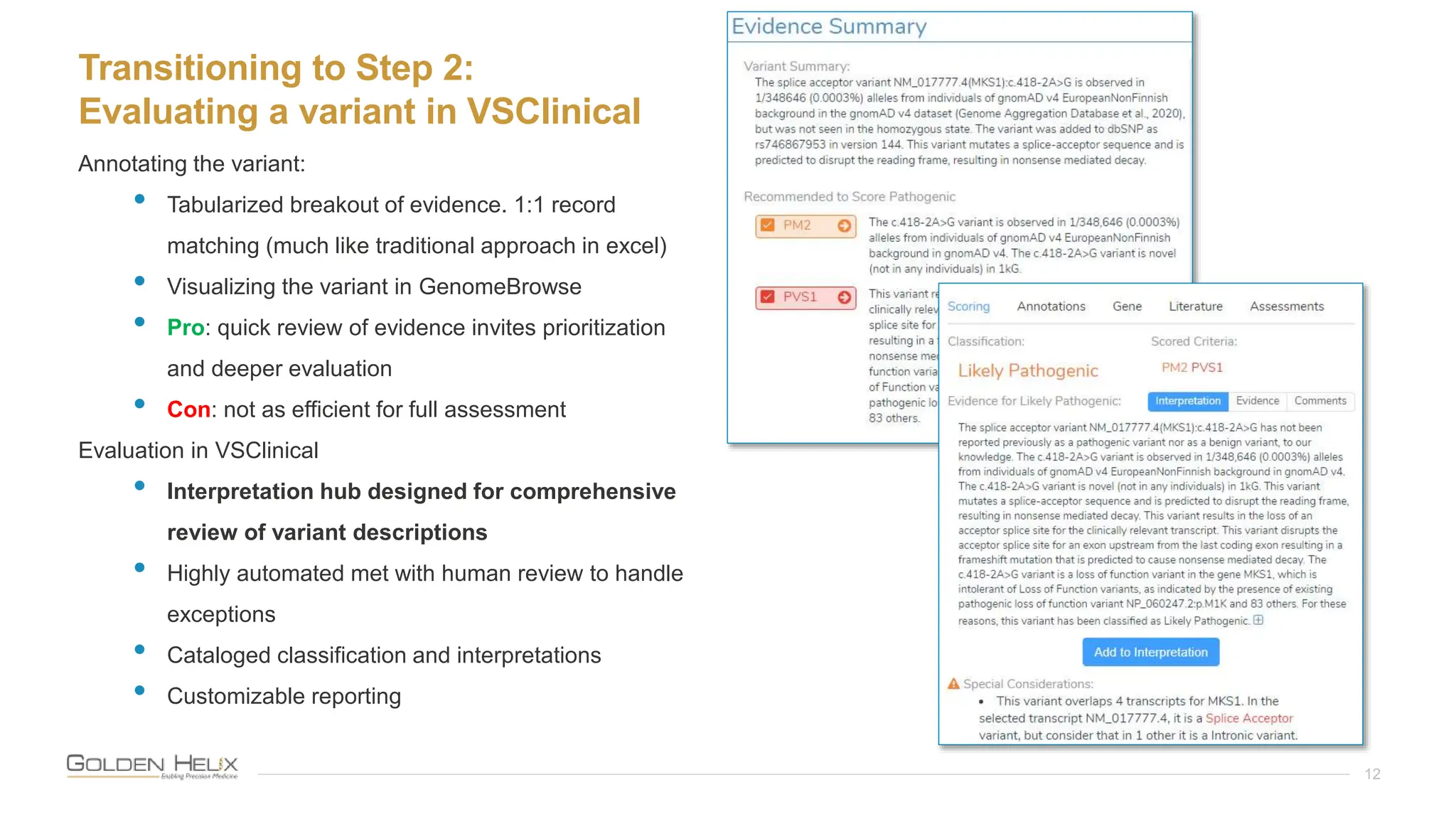
The document details a presentation by Darby Kammeraad on the complete variant assessment in VsClinical, supported by NIH grant funding and highlighting the software's capabilities in genomic analysis for various clinical applications. Golden Helix, founded in 1998, offers a SaaS bioinformatics solution for next-gen sequencing data analysis, with a focus on quality management and certifications. The presentation also discusses recent webcasts, the filtering and evaluation of genetic variants, and the clinical benefits of their software offerings.