Fit for fitting: Bionext-DSS, a dataset of similars sites
•
0 likes•138 views
“Bionext-DSS” is a dataset of similar binding sites which contains 35 test cases connected to a pharmaceutical application. Each case includes a reference site, as well as several positive sites further classified in 6 bins according to their similarity level to the reference. We moreover provide a list of noise structures carefully selected so as to be used as negative controls in algorithms evaluation.
Report
Share
Report
Share
Download to read offline
Recommended
Pharmacohoreppt

1) Pharmacophores are sets of steric and electronic features common to active drug molecules that interact with biological targets in a specific way. They include features like hydrogen bond donors/acceptors and hydrophobic regions.
2) Feature trees (Ftrees) are a ligand-based approach that represents molecules as trees to capture major building blocks and overall alignment in a conformation-independent way, supporting "lead hopping" between chemical classes.
3) Ftrees describe molecular fragments as nodes labeled with shape and chemical descriptors. Molecules are compared by matching subtrees using topology-preserving search algorithms. This allows identification of actives from different chemical scaffolds.
Cook2010web

The document provides an overview of computational chemistry methods for structure-activity relationship analysis, pharmacophore modeling, and protein-ligand docking. It discusses topics like SAR, QSAR, molecular alignment, conformational analysis, homology modeling of protein targets, and docking programs. Examples are given of applying these methods to study benzodiazepine ligands and GABA receptor subtypes.
Molecular docking by harendra ...power point presentation

Molecular docking is a computational method used to predict how a small molecule, like a drug, binds to a larger target molecule, like a protein. It works by fitting the structures of the two molecules together to find the highest affinity binding mode. The docking process involves defining the active site on the target protein, generating possible positions for the small molecule to bind, scoring the interactions between them, and identifying the best binding pose. Docking can help researchers design new drugs that effectively interact with protein targets.
consensus superiority of the pharmacophore based alignment, over maximum comm...

The document discusses 3D-QSAR studies conducted on a set of 52 carbamate compounds with acetylcholinesterase inhibitory activity. The compounds were divided into training and test sets. Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) models were developed using two alignment strategies: Maximum Common Substructure (MCS)-based alignment and pharmacophore-based alignment from Hip-Hop algorithm. The pharmacophore-based alignment produced superior models with predictive r2 values of 0.614 and 0.788 for CoMFA and CoMSIA respectively. Steric effects and hydrophobicity were found to play important roles in inhibitory activity. The studies suggest pharmac
Beatson DDU Fragment Lbrary Design Poster

1) The researchers redesigned their fragment library to enhance chemical diversity and 3D content by combining commercial and in-house databases.
2) They analyzed shape, chemical, and pharmacophore diversity of the new library and found it had greater diversity than the original library and a commercial diversity library while maintaining good physicochemical properties and lower complexity.
3) Future plans include synthesizing fragments from biologically active molecules and enumerated scaffolds to further increase diversity of the library.
Dispensing Processes Impact Apparent Biological Activity as Determined by Com...

Dispensing Processes Impact Apparent Biological Activity as Determined by Com...US Environmental Protection Agency (EPA), Center for Computational Toxicology and Exposure
Dispensing and dilution processes may profoundly influence estimates of biological activity of compounds. Published data show Ephrin type-B receptor 4 IC50 values obtained via tip-based serial dilution and dispensing versus acoustic dispensing with direct dilution differ by orders of magnitude with no correlation or ranking of datasets. We generated computational 3D pharmacophores based on data derived by both acoustic and tip-based transfer. The computed pharmacophores differ significantly depending upon dispensing and dilution methods. The acoustic dispensing-derived pharmacophore correctly identified active compounds in a subsequent test set where the tip-based method failed. Data from acoustic dispensing generates a pharmacophore containing two hydrophobic features, one hydrogen bond donor and one hydrogen bond acceptor. This is consistent with X-ray crystallography studies of ligand-protein interactions and automatically generated pharmacophores derived from this structural data. In contrast, the tip-based data suggest a pharmacophore with two hydrogen bond acceptors, one hydrogen bond donor and no hydrophobic features. This pharmacophore is inconsistent with the X-ray crystallographic studies and automatically generated pharmacophores. In short, traditional dispensing processes are another important source of error in high-throughput screening that impacts computational and statistical analyses. These findings have far-reaching implications in biological research.3d qsar

3D QSAR techniques like CoMFA and CoMSIA can quantitatively correlate the biological activity of a series of compounds to their 3D molecular properties. CoMFA generates 3D interaction fields around aligned molecules using steric and electrostatic probes, while CoMSIA additionally considers hydrophobic and hydrogen bonding interactions. The document discusses these techniques and provides an example case study applying CoMFA to develop a QSAR model for human eosinophil phosphodiesterase inhibitors with a cross-validated R2 of 0.565. In conclusion, 3D QSAR is a valuable tool for understanding structure-activity relationships and aiding drug design and discovery efforts.
Computer Aided Drug Design

Computer aided drug design uses computational approaches to aid in the drug discovery process. There are several key approaches including ligand based approaches which identify characteristics of known active ligands, target based approaches which use information about the biological target, and structure based drug design which utilizes 3D structural information. The main steps in drug design include target identification and validation, lead identification and optimization, and preclinical and clinical trials. Computational tools are used throughout the process for tasks like molecular docking, ADMET prediction, and structure activity relationship analysis.
Recommended
Pharmacohoreppt

1) Pharmacophores are sets of steric and electronic features common to active drug molecules that interact with biological targets in a specific way. They include features like hydrogen bond donors/acceptors and hydrophobic regions.
2) Feature trees (Ftrees) are a ligand-based approach that represents molecules as trees to capture major building blocks and overall alignment in a conformation-independent way, supporting "lead hopping" between chemical classes.
3) Ftrees describe molecular fragments as nodes labeled with shape and chemical descriptors. Molecules are compared by matching subtrees using topology-preserving search algorithms. This allows identification of actives from different chemical scaffolds.
Cook2010web

The document provides an overview of computational chemistry methods for structure-activity relationship analysis, pharmacophore modeling, and protein-ligand docking. It discusses topics like SAR, QSAR, molecular alignment, conformational analysis, homology modeling of protein targets, and docking programs. Examples are given of applying these methods to study benzodiazepine ligands and GABA receptor subtypes.
Molecular docking by harendra ...power point presentation

Molecular docking is a computational method used to predict how a small molecule, like a drug, binds to a larger target molecule, like a protein. It works by fitting the structures of the two molecules together to find the highest affinity binding mode. The docking process involves defining the active site on the target protein, generating possible positions for the small molecule to bind, scoring the interactions between them, and identifying the best binding pose. Docking can help researchers design new drugs that effectively interact with protein targets.
consensus superiority of the pharmacophore based alignment, over maximum comm...

The document discusses 3D-QSAR studies conducted on a set of 52 carbamate compounds with acetylcholinesterase inhibitory activity. The compounds were divided into training and test sets. Comparative Molecular Field Analysis (CoMFA) and Comparative Molecular Similarity Indices Analysis (CoMSIA) models were developed using two alignment strategies: Maximum Common Substructure (MCS)-based alignment and pharmacophore-based alignment from Hip-Hop algorithm. The pharmacophore-based alignment produced superior models with predictive r2 values of 0.614 and 0.788 for CoMFA and CoMSIA respectively. Steric effects and hydrophobicity were found to play important roles in inhibitory activity. The studies suggest pharmac
Beatson DDU Fragment Lbrary Design Poster

1) The researchers redesigned their fragment library to enhance chemical diversity and 3D content by combining commercial and in-house databases.
2) They analyzed shape, chemical, and pharmacophore diversity of the new library and found it had greater diversity than the original library and a commercial diversity library while maintaining good physicochemical properties and lower complexity.
3) Future plans include synthesizing fragments from biologically active molecules and enumerated scaffolds to further increase diversity of the library.
Dispensing Processes Impact Apparent Biological Activity as Determined by Com...

Dispensing Processes Impact Apparent Biological Activity as Determined by Com...US Environmental Protection Agency (EPA), Center for Computational Toxicology and Exposure
Dispensing and dilution processes may profoundly influence estimates of biological activity of compounds. Published data show Ephrin type-B receptor 4 IC50 values obtained via tip-based serial dilution and dispensing versus acoustic dispensing with direct dilution differ by orders of magnitude with no correlation or ranking of datasets. We generated computational 3D pharmacophores based on data derived by both acoustic and tip-based transfer. The computed pharmacophores differ significantly depending upon dispensing and dilution methods. The acoustic dispensing-derived pharmacophore correctly identified active compounds in a subsequent test set where the tip-based method failed. Data from acoustic dispensing generates a pharmacophore containing two hydrophobic features, one hydrogen bond donor and one hydrogen bond acceptor. This is consistent with X-ray crystallography studies of ligand-protein interactions and automatically generated pharmacophores derived from this structural data. In contrast, the tip-based data suggest a pharmacophore with two hydrogen bond acceptors, one hydrogen bond donor and no hydrophobic features. This pharmacophore is inconsistent with the X-ray crystallographic studies and automatically generated pharmacophores. In short, traditional dispensing processes are another important source of error in high-throughput screening that impacts computational and statistical analyses. These findings have far-reaching implications in biological research.3d qsar

3D QSAR techniques like CoMFA and CoMSIA can quantitatively correlate the biological activity of a series of compounds to their 3D molecular properties. CoMFA generates 3D interaction fields around aligned molecules using steric and electrostatic probes, while CoMSIA additionally considers hydrophobic and hydrogen bonding interactions. The document discusses these techniques and provides an example case study applying CoMFA to develop a QSAR model for human eosinophil phosphodiesterase inhibitors with a cross-validated R2 of 0.565. In conclusion, 3D QSAR is a valuable tool for understanding structure-activity relationships and aiding drug design and discovery efforts.
Computer Aided Drug Design

Computer aided drug design uses computational approaches to aid in the drug discovery process. There are several key approaches including ligand based approaches which identify characteristics of known active ligands, target based approaches which use information about the biological target, and structure based drug design which utilizes 3D structural information. The main steps in drug design include target identification and validation, lead identification and optimization, and preclinical and clinical trials. Computational tools are used throughout the process for tasks like molecular docking, ADMET prediction, and structure activity relationship analysis.
Target oriented generic fingerprint-based molecular representation

The screening of chemical libraries is an important step in the drug discovery process. The
existing chemical libraries contain up to millions of compounds. As the screening at such scale
is expensive, the virtual screening is often utilized. There exist several variants of virtual
screening and ligand-based virtual screening is one of them. It utilizes the similarity of screened
chemical compounds to known compounds. Besides the employed similarity measure, another
aspect greatly influencing the performance of ligand-based virtual screening is the chosen
chemical compound representation. In this paper, we introduce a fragment-based
representation of chemical compounds. Our representation utilizes fragments to represent a
compound where each fragment is represented by its physico-chemical descriptors. The
representation is highly parametrizable, especially in the area of physico-chemical descriptors
selection and application. In order to test the performance of our method, we utilized an existing
framework for virtual screening benchmarking. The results show that our method is comparable
to the best existing approaches and on some data sets it outperforms them.
Computational Drug Design

Computational drug design uses computer-aided drug design (CADD) approaches like structure-based drug design, ligand-based drug design, and protein-ligand docking to rationally design new drug candidates. These CADD approaches leverage information about protein and ligand structures to predict how well potential drug molecules may bind to their target without relying on experimental screening. Key techniques include pharmacophore modeling, virtual screening, and predicting the binding pose and affinity of ligands docked in the target protein's active site. Accurate preparation of the protein structure is important for successful structure-based drug design applications like protein-ligand docking.
Ligand based drug design

1. Quantitative Structure Activity Relationship (QSAR) uses mathematical models to correlate chemical descriptors of molecules to their biological activity.
2. Descriptors include physicochemical properties like hydrophobicity which can be measured experimentally.
3. QSAR allows medicinal chemists to predict activity of novel analogues and guide synthesis toward more active compounds.
In Silico methods for ADMET prediction of new molecules

The document discusses the importance of predicting absorption, distribution, metabolism, excretion and toxicity (ADMET) properties of new molecules in silico during drug design. It describes how ADMET prediction techniques have evolved since 1863 and helped advance drug development. Factors considered in developing ADMET prediction models include the model purpose, required prediction speed and accuracy. Common molecular descriptors used in these models are also discussed. The document outlines methods for predicting various ADMET properties like permeability, solubility, distribution and metabolism in silico. Recent tools for computational ADMET prediction are also mentioned.
Structure based and ligand based drug designing

This document discusses structure-based and ligand-based drug design approaches. Structure-based design uses the 3D structure of biological targets to dock potential drug molecules. Ligand-based design analyzes similar molecules that bind to the target to derive pharmacophore models or quantitative structure-activity relationships (QSAR) to predict new candidates. Specific structure-based methods covered include docking tools like AutoDock and CDOCKER, and accounting for protein and complex flexibility. Ligand-based methods discussed are QSAR techniques like Comparative Molecular Field Analysis (CoMSIA) and Field Analysis (CoMFA). In conclusion, computational approaches like these are valuable for drug discovery by facilitating the identification and testing of new ligand
Structure based drug design- kiranmayi

This presentation helps in detail learning about the structure based drug design. It includes types of structure based drug design and detailed study of docking, de novo drug design.
Validation of Clomipramine interactions identified by BioBind against experim...

Based on the now accepted principle that
similar receptors bind similars ligands, we have developed BioBind, a patented comparison algorithm
dedicated to the retrieval and assessment of local surface similarities. Clomipramine appears to be a
good “real life” candidate to challenge BioBind. In a couple of hours, BioBind was able to retrieve all
known targets having structural data described in the literature and to provide a valuable list of unknown
yet sensible putative targets currently being experimentally validated. This analysis hence demonstrates the
robustness and relevance of BioBind.
Chemistry Reserach as a Social Machine

Chemistry reserarch is undergoing signricant changes with the increasing use of digital tools and artifical and agumented intelegence
ENHANCED POPULATION BASED ANT COLONY FOR THE 3D HYDROPHOBIC POLAR PROTEIN STR...

Population-based Ant Colony algorithm is stochastic local search algorithm that mimics the behavior of
real ants, simulating pheromone trails to search for solutions to combinatorial optimization problems. This
paper introduces population-based Ant Colony algorithm to solve 3D Hydrophobic Polar Protein structure
Prediction Problem then introduces a new enhanced approach of population-based Ant Colony algorithm
called Enhanced Population-based Ant Colony algorithm (EP-ACO) to avoid stagnation problem in
population-based Ant Colony algorithm and increase exploration in the search space escaping from local
optima, The experiments show that our approach appears more efficient results than state of art method.
Pep Talk San Diego 011311

Presentation made at PepTalk 2011 in San Diego on Jan. 13, 2011. The emphasis is on computational methods to explore global and local structure similarities in determining the possible promiscuity of drugs to bind to multiple protein receptors.
Homology modeling

Homology modeling is a technique used to predict the 3D structure of a protein based on the alignment of its amino acid sequence to known protein structures. It relies on the observation that structure is more conserved than sequence during evolution. The key steps in homology modeling include: 1) identifying a template structure through sequence alignment tools like BLAST, 2) correcting any errors in the initial alignment, 3) generating the protein backbone based on the template structure, 4) modeling any loops or missing regions, 5) adding side chains, 6) optimizing the model structure energetically, and 7) validating that the final model matches the template structure and has correct stereochemistry. Homology modeling is useful for applications like structure-based drug design
PaulZRXie_Nucl. Acids Res.-2013

This document summarizes a web server called LISE that predicts ligand-binding sites on proteins. It does so using a novel method that analyzes geometric motifs called "protein triangles" extracted from interaction networks between protein and ligand atoms. The server has been thoroughly evaluated on a large dataset of over 2000 protein structures, representing the largest test ever conducted for a ligand-binding site prediction method. The server is freely available online and automatically predicts binding sites for over 63,000 protein structures from the PDB, or can perform new predictions on user-submitted structures. Evaluation on several benchmark datasets demonstrates it achieves success rates over 70% for top-ranked predictions and over 85% for top-3 predictions.
E0362430

The document describes a molecular docking study of aspirin and aspirin derivatives using the HVR protein (HIV protease receptor). The study found that compounds SR-03, SR-02, and SR-04 showed the best docking scores and interactions with the HVR protein, indicating they may have potential anti-HIV activity. It was concluded that electron withdrawing groups attached to the aryl substituent of the carboxylic acid group in aspirin increase affinity for the HIV protein, while electron donating groups decrease affinity. Further studies are needed to determine the exact mechanisms of action.
Determining stable ligand orientation

Each and every biological function in living organism occurs due to protein-protein interactions. The
diseases are no exception to this. Identifying one or more proteins for a particular disease and then
designing a suitable chemical compound (which is known as drug or ligand) to destroy those proteins is a
challenging topic of research in computational biology. In earlier methods, drugs were designed using only
a few chemical components and were represented as a fixed-length tree. But in reality, a drug contains
many chemical groups collectively known as pharmacophore. Moreover, the chemical length of the drug
cannot be determined before designing that drug.
In the present work, a Particle Swarm Optimization (PSO) based methodology has been proposed to find
out a suitable drug for a particular disease so that the drug-target protein interaction energy becomes
minimum. In the proposed algorithm, the drug is represented as a variable length tree and essential
functional groups are arranged in different positions of that drug. Finally, the structure of the drug is
obtained and its docking energy is minimized simultaneously. Also, the orientation of chemical groups in
the drug is tested so that it can bind to a particular active site of a target protein and the drug fits well
inside the active site of target protein. Here, several inter-molecular forces have been considered for
accuracy of the docking energy. Results are demonstrated for three different target proteins both
numerically and pictorially. Results show that PSO performs better than the earlier methods.
Cadd and molecular modeling for M.Pharm

THE CADD IS FOR THE DRUG DEVELOPMENT THE DIFFERENT STRATEGIES ARE MENTIONED LIKE QSAR MOLECULAR DOCKING, THE DIFFERENT DIMNSIONAL FORMS OF QSAR , THE ADVANCE SAR of it.
HMM’S INTERPOLATION OF PROTIENS FOR PROFILE ANALYSIS

HMM has found its application in almost every field. Applying Hmm to biological sequences has its own
advantages. HMM’s being more systematic and specific, yield a result better than consensus techniques.
Profile HMMs use position specific scoring for the matching & substitution of a residue and for the
opening or extension of a gap. HMMs apply a statistical method to estimate the true frequency of a residue
at a given position in the alignment from its observed frequency while standard profiles use the observed
frequency itself to assign the score for that residue. This means that a profile HMM derived from only 10 to
20 aligned sequences can be of equivalent quality to a standard profile created from 40 to 50 aligned
sequences.
Stable Drug Designing by Minimizing Drug Protein Interaction Energy Using PSO 

1. The document proposes using a particle swarm optimization (PSO) algorithm to design stable drug molecules that minimize interaction energy with target proteins.
2. In the algorithm, drugs are represented as variable-length trees containing functional groups, and PSO is used to optimize van der Waals and electrostatic interaction energies.
3. Results show that PSO performs better than previous fixed-length tree methods at designing drugs that stably bind to active sites of human rhinovirus, malaria, and HIV proteins.
Bind database

The document discusses the Biomolecular Interaction Network Database (BIND), which stores information about molecular interactions, complexes, and pathways. BIND uses standards like ASN.1 and XML to specify interactions. It stores details about molecules, interactions, publications, and more. Tools like Pajek and MCODE are used to visualize and analyze the network. The database has expanded to include additional details like post-translational modifications, cellular localization, and links to other databases. Manual and automatic submission is supported.
BCSRCv1.3

The document describes a computational study aimed at expediting drug discovery by identifying novel protein-protein interaction (PPI) ligands. The study used computational chemistry programs to test ligands against protein structures and identify those that overlay protein secondary structures with a root-mean-square deviation (RMSD) below 0.5 angstroms. Many ligands were found to successfully mimic protein secondary structures with low RMSD values, supporting the hypothesis that novel PPI ligands can be identified in this manner. The results indicate this computational approach may speed up drug discovery by targeting PPIs rather than single protein inhibition.
Research Inventy : International Journal of Engineering and Science

Research Inventy : International Journal of Engineering and Science is published by the group of young academic and industrial researchers with 12 Issues per year. It is an online as well as print version open access journal that provides rapid publication (monthly) of articles in all areas of the subject such as: civil, mechanical, chemical, electronic and computer engineering as well as production and information technology. The Journal welcomes the submission of manuscripts that meet the general criteria of significance and scientific excellence. Papers will be published by rapid process within 20 days after acceptance and peer review process takes only 7 days. All articles published in Research Inventy will be peer-reviewed.
MULISA : A New Strategy for Discovery of Protein Functional Motifs and Residues

To predict and identify details regarding function
from protein sequences is an emergency task
since the growing number and diversity of protein s
equence. Here, we develop a novel approach
for identifying conservation residues and motifs of
ligand-binding proteins. In this method,
called MuLiSA (Multiple Ligand-bound Structure Alig
nment), we first superimpose the ligands
of ligand-binding proteins and then the residues of
ligand-binding sites are naturally aligned.
We identify important residues and patterns based o
n the z-scores of the residue entropy and
residue-segment entropy. After identifying new patt
ern candidates, the profiles of patterns are
generated to predict the protein function from only
protein sequences. We tested our approach
on ATP-binding proteins and HEM-binding proteins. T
he experiments show that MuLiSA can
identify the conservation residues and novel patter
ns which are really correlated with protein
functions of certain ligand-binding proteins. We fo
und that our MuLiSA can identify
conservation patterns and is better than traditiona
l alignments such as CE and CLUSTALW in
some ligand-binding proteins. We believe that our M
uLiSA is useful to discover ligand-binding
specificity-determining residues and functional imp
ortant patterns of proteins.
RATIONAL DRUG DESIGN.pptx

This document discusses computer-aided drug design (CADD) and the use of pharmacophore modeling to aid in the drug discovery process. It provides details on the basic principles of developing a 3D pharmacophore model, including identifying active conformations of ligands that overlap on common pharmacophore elements. The selection of appropriate pharmacophore elements from a set of active ligands is also covered. Methods for generating pharmacophore models, both manually and automatically, are described.
More Related Content
What's hot
Target oriented generic fingerprint-based molecular representation

The screening of chemical libraries is an important step in the drug discovery process. The
existing chemical libraries contain up to millions of compounds. As the screening at such scale
is expensive, the virtual screening is often utilized. There exist several variants of virtual
screening and ligand-based virtual screening is one of them. It utilizes the similarity of screened
chemical compounds to known compounds. Besides the employed similarity measure, another
aspect greatly influencing the performance of ligand-based virtual screening is the chosen
chemical compound representation. In this paper, we introduce a fragment-based
representation of chemical compounds. Our representation utilizes fragments to represent a
compound where each fragment is represented by its physico-chemical descriptors. The
representation is highly parametrizable, especially in the area of physico-chemical descriptors
selection and application. In order to test the performance of our method, we utilized an existing
framework for virtual screening benchmarking. The results show that our method is comparable
to the best existing approaches and on some data sets it outperforms them.
Computational Drug Design

Computational drug design uses computer-aided drug design (CADD) approaches like structure-based drug design, ligand-based drug design, and protein-ligand docking to rationally design new drug candidates. These CADD approaches leverage information about protein and ligand structures to predict how well potential drug molecules may bind to their target without relying on experimental screening. Key techniques include pharmacophore modeling, virtual screening, and predicting the binding pose and affinity of ligands docked in the target protein's active site. Accurate preparation of the protein structure is important for successful structure-based drug design applications like protein-ligand docking.
Ligand based drug design

1. Quantitative Structure Activity Relationship (QSAR) uses mathematical models to correlate chemical descriptors of molecules to their biological activity.
2. Descriptors include physicochemical properties like hydrophobicity which can be measured experimentally.
3. QSAR allows medicinal chemists to predict activity of novel analogues and guide synthesis toward more active compounds.
In Silico methods for ADMET prediction of new molecules

The document discusses the importance of predicting absorption, distribution, metabolism, excretion and toxicity (ADMET) properties of new molecules in silico during drug design. It describes how ADMET prediction techniques have evolved since 1863 and helped advance drug development. Factors considered in developing ADMET prediction models include the model purpose, required prediction speed and accuracy. Common molecular descriptors used in these models are also discussed. The document outlines methods for predicting various ADMET properties like permeability, solubility, distribution and metabolism in silico. Recent tools for computational ADMET prediction are also mentioned.
Structure based and ligand based drug designing

This document discusses structure-based and ligand-based drug design approaches. Structure-based design uses the 3D structure of biological targets to dock potential drug molecules. Ligand-based design analyzes similar molecules that bind to the target to derive pharmacophore models or quantitative structure-activity relationships (QSAR) to predict new candidates. Specific structure-based methods covered include docking tools like AutoDock and CDOCKER, and accounting for protein and complex flexibility. Ligand-based methods discussed are QSAR techniques like Comparative Molecular Field Analysis (CoMSIA) and Field Analysis (CoMFA). In conclusion, computational approaches like these are valuable for drug discovery by facilitating the identification and testing of new ligand
Structure based drug design- kiranmayi

This presentation helps in detail learning about the structure based drug design. It includes types of structure based drug design and detailed study of docking, de novo drug design.
What's hot (6)
Target oriented generic fingerprint-based molecular representation

Target oriented generic fingerprint-based molecular representation
In Silico methods for ADMET prediction of new molecules

In Silico methods for ADMET prediction of new molecules
Similar to Fit for fitting: Bionext-DSS, a dataset of similars sites
Validation of Clomipramine interactions identified by BioBind against experim...

Based on the now accepted principle that
similar receptors bind similars ligands, we have developed BioBind, a patented comparison algorithm
dedicated to the retrieval and assessment of local surface similarities. Clomipramine appears to be a
good “real life” candidate to challenge BioBind. In a couple of hours, BioBind was able to retrieve all
known targets having structural data described in the literature and to provide a valuable list of unknown
yet sensible putative targets currently being experimentally validated. This analysis hence demonstrates the
robustness and relevance of BioBind.
Chemistry Reserach as a Social Machine

Chemistry reserarch is undergoing signricant changes with the increasing use of digital tools and artifical and agumented intelegence
ENHANCED POPULATION BASED ANT COLONY FOR THE 3D HYDROPHOBIC POLAR PROTEIN STR...

Population-based Ant Colony algorithm is stochastic local search algorithm that mimics the behavior of
real ants, simulating pheromone trails to search for solutions to combinatorial optimization problems. This
paper introduces population-based Ant Colony algorithm to solve 3D Hydrophobic Polar Protein structure
Prediction Problem then introduces a new enhanced approach of population-based Ant Colony algorithm
called Enhanced Population-based Ant Colony algorithm (EP-ACO) to avoid stagnation problem in
population-based Ant Colony algorithm and increase exploration in the search space escaping from local
optima, The experiments show that our approach appears more efficient results than state of art method.
Pep Talk San Diego 011311

Presentation made at PepTalk 2011 in San Diego on Jan. 13, 2011. The emphasis is on computational methods to explore global and local structure similarities in determining the possible promiscuity of drugs to bind to multiple protein receptors.
Homology modeling

Homology modeling is a technique used to predict the 3D structure of a protein based on the alignment of its amino acid sequence to known protein structures. It relies on the observation that structure is more conserved than sequence during evolution. The key steps in homology modeling include: 1) identifying a template structure through sequence alignment tools like BLAST, 2) correcting any errors in the initial alignment, 3) generating the protein backbone based on the template structure, 4) modeling any loops or missing regions, 5) adding side chains, 6) optimizing the model structure energetically, and 7) validating that the final model matches the template structure and has correct stereochemistry. Homology modeling is useful for applications like structure-based drug design
PaulZRXie_Nucl. Acids Res.-2013

This document summarizes a web server called LISE that predicts ligand-binding sites on proteins. It does so using a novel method that analyzes geometric motifs called "protein triangles" extracted from interaction networks between protein and ligand atoms. The server has been thoroughly evaluated on a large dataset of over 2000 protein structures, representing the largest test ever conducted for a ligand-binding site prediction method. The server is freely available online and automatically predicts binding sites for over 63,000 protein structures from the PDB, or can perform new predictions on user-submitted structures. Evaluation on several benchmark datasets demonstrates it achieves success rates over 70% for top-ranked predictions and over 85% for top-3 predictions.
E0362430

The document describes a molecular docking study of aspirin and aspirin derivatives using the HVR protein (HIV protease receptor). The study found that compounds SR-03, SR-02, and SR-04 showed the best docking scores and interactions with the HVR protein, indicating they may have potential anti-HIV activity. It was concluded that electron withdrawing groups attached to the aryl substituent of the carboxylic acid group in aspirin increase affinity for the HIV protein, while electron donating groups decrease affinity. Further studies are needed to determine the exact mechanisms of action.
Determining stable ligand orientation

Each and every biological function in living organism occurs due to protein-protein interactions. The
diseases are no exception to this. Identifying one or more proteins for a particular disease and then
designing a suitable chemical compound (which is known as drug or ligand) to destroy those proteins is a
challenging topic of research in computational biology. In earlier methods, drugs were designed using only
a few chemical components and were represented as a fixed-length tree. But in reality, a drug contains
many chemical groups collectively known as pharmacophore. Moreover, the chemical length of the drug
cannot be determined before designing that drug.
In the present work, a Particle Swarm Optimization (PSO) based methodology has been proposed to find
out a suitable drug for a particular disease so that the drug-target protein interaction energy becomes
minimum. In the proposed algorithm, the drug is represented as a variable length tree and essential
functional groups are arranged in different positions of that drug. Finally, the structure of the drug is
obtained and its docking energy is minimized simultaneously. Also, the orientation of chemical groups in
the drug is tested so that it can bind to a particular active site of a target protein and the drug fits well
inside the active site of target protein. Here, several inter-molecular forces have been considered for
accuracy of the docking energy. Results are demonstrated for three different target proteins both
numerically and pictorially. Results show that PSO performs better than the earlier methods.
Cadd and molecular modeling for M.Pharm

THE CADD IS FOR THE DRUG DEVELOPMENT THE DIFFERENT STRATEGIES ARE MENTIONED LIKE QSAR MOLECULAR DOCKING, THE DIFFERENT DIMNSIONAL FORMS OF QSAR , THE ADVANCE SAR of it.
HMM’S INTERPOLATION OF PROTIENS FOR PROFILE ANALYSIS

HMM has found its application in almost every field. Applying Hmm to biological sequences has its own
advantages. HMM’s being more systematic and specific, yield a result better than consensus techniques.
Profile HMMs use position specific scoring for the matching & substitution of a residue and for the
opening or extension of a gap. HMMs apply a statistical method to estimate the true frequency of a residue
at a given position in the alignment from its observed frequency while standard profiles use the observed
frequency itself to assign the score for that residue. This means that a profile HMM derived from only 10 to
20 aligned sequences can be of equivalent quality to a standard profile created from 40 to 50 aligned
sequences.
Stable Drug Designing by Minimizing Drug Protein Interaction Energy Using PSO 

1. The document proposes using a particle swarm optimization (PSO) algorithm to design stable drug molecules that minimize interaction energy with target proteins.
2. In the algorithm, drugs are represented as variable-length trees containing functional groups, and PSO is used to optimize van der Waals and electrostatic interaction energies.
3. Results show that PSO performs better than previous fixed-length tree methods at designing drugs that stably bind to active sites of human rhinovirus, malaria, and HIV proteins.
Bind database

The document discusses the Biomolecular Interaction Network Database (BIND), which stores information about molecular interactions, complexes, and pathways. BIND uses standards like ASN.1 and XML to specify interactions. It stores details about molecules, interactions, publications, and more. Tools like Pajek and MCODE are used to visualize and analyze the network. The database has expanded to include additional details like post-translational modifications, cellular localization, and links to other databases. Manual and automatic submission is supported.
BCSRCv1.3

The document describes a computational study aimed at expediting drug discovery by identifying novel protein-protein interaction (PPI) ligands. The study used computational chemistry programs to test ligands against protein structures and identify those that overlay protein secondary structures with a root-mean-square deviation (RMSD) below 0.5 angstroms. Many ligands were found to successfully mimic protein secondary structures with low RMSD values, supporting the hypothesis that novel PPI ligands can be identified in this manner. The results indicate this computational approach may speed up drug discovery by targeting PPIs rather than single protein inhibition.
Research Inventy : International Journal of Engineering and Science

Research Inventy : International Journal of Engineering and Science is published by the group of young academic and industrial researchers with 12 Issues per year. It is an online as well as print version open access journal that provides rapid publication (monthly) of articles in all areas of the subject such as: civil, mechanical, chemical, electronic and computer engineering as well as production and information technology. The Journal welcomes the submission of manuscripts that meet the general criteria of significance and scientific excellence. Papers will be published by rapid process within 20 days after acceptance and peer review process takes only 7 days. All articles published in Research Inventy will be peer-reviewed.
MULISA : A New Strategy for Discovery of Protein Functional Motifs and Residues

To predict and identify details regarding function
from protein sequences is an emergency task
since the growing number and diversity of protein s
equence. Here, we develop a novel approach
for identifying conservation residues and motifs of
ligand-binding proteins. In this method,
called MuLiSA (Multiple Ligand-bound Structure Alig
nment), we first superimpose the ligands
of ligand-binding proteins and then the residues of
ligand-binding sites are naturally aligned.
We identify important residues and patterns based o
n the z-scores of the residue entropy and
residue-segment entropy. After identifying new patt
ern candidates, the profiles of patterns are
generated to predict the protein function from only
protein sequences. We tested our approach
on ATP-binding proteins and HEM-binding proteins. T
he experiments show that MuLiSA can
identify the conservation residues and novel patter
ns which are really correlated with protein
functions of certain ligand-binding proteins. We fo
und that our MuLiSA can identify
conservation patterns and is better than traditiona
l alignments such as CE and CLUSTALW in
some ligand-binding proteins. We believe that our M
uLiSA is useful to discover ligand-binding
specificity-determining residues and functional imp
ortant patterns of proteins.
RATIONAL DRUG DESIGN.pptx

This document discusses computer-aided drug design (CADD) and the use of pharmacophore modeling to aid in the drug discovery process. It provides details on the basic principles of developing a 3D pharmacophore model, including identifying active conformations of ligands that overlap on common pharmacophore elements. The selection of appropriate pharmacophore elements from a set of active ligands is also covered. Methods for generating pharmacophore models, both manually and automatically, are described.
Structure based drug designing

This document discusses structure based drug design. It describes how drug design uses knowledge of biological targets to find new medications. Structure based drug design uses information about the 3D structure of protein targets to design ligands that bind to them. The main methods described are ligand-based drug design through database searching, and receptor-based drug design which builds ligands for a receptor. Molecular docking is also discussed as a key technique to predict how ligands bind to protein targets and identify potential drug candidates.
Cadd assignment 4 (sarita)

The document discusses various topics related to computer-aided drug design (CADD), including:
1) The definitions of drug-likeness, druggability, and the Rule of Five for screening drug-like molecules. The Rule of Five outlines molecular properties important for a drug's absorption and metabolism.
2) Pharmacophore-based and ligand-based virtual screening methods which use the structure of known active ligands to search compound libraries for similar molecules.
3) The role of virtual screening in CADD to select compounds for biological testing from large databases using techniques like structure-based docking and ligand-based similarity searching. Scoring functions are also used to rank compounds.
Ppi

This document summarizes a presentation on protein-protein interactions. It discusses biological aspects of PPIs and introduces several PPI databases and tools. The presentation is divided into sections on the introduction of PPIs, databases like BIND and DIP, pathways and algorithms, and visualization tools. It provides information on the types and methods of studying PPIs experimentally.
Threading modeling methods

This document discusses protein threading modeling methods. Protein threading, also called fold recognition, is used to model proteins that have the same fold as proteins with known structures but no homologous sequences. It differs from homology modeling which is used for proteins that have homologous sequences. Protein threading works by using statistical knowledge of relationships between structures in the Protein Data Bank and the sequence of the protein being modeled. It is based on observations that there are a limited number of folds in nature and most new structures have similar folds to ones already in the PDB. The document then describes the general steps of the protein threading method.
Similar to Fit for fitting: Bionext-DSS, a dataset of similars sites (20)
Validation of Clomipramine interactions identified by BioBind against experim...

Validation of Clomipramine interactions identified by BioBind against experim...
ENHANCED POPULATION BASED ANT COLONY FOR THE 3D HYDROPHOBIC POLAR PROTEIN STR...

ENHANCED POPULATION BASED ANT COLONY FOR THE 3D HYDROPHOBIC POLAR PROTEIN STR...
HMM’S INTERPOLATION OF PROTIENS FOR PROFILE ANALYSIS

HMM’S INTERPOLATION OF PROTIENS FOR PROFILE ANALYSIS
Stable Drug Designing by Minimizing Drug Protein Interaction Energy Using PSO 

Stable Drug Designing by Minimizing Drug Protein Interaction Energy Using PSO
Research Inventy : International Journal of Engineering and Science

Research Inventy : International Journal of Engineering and Science
MULISA : A New Strategy for Discovery of Protein Functional Motifs and Residues

MULISA : A New Strategy for Discovery of Protein Functional Motifs and Residues
Recently uploaded
Role of Mukta Pishti in the Management of Hyperthyroidism

Muktapishti is a traditional Ayurvedic preparation made from Shoditha Mukta (Purified Pearl), is believed to help regulate thyroid function and reduce symptoms of hyperthyroidism due to its cooling and balancing properties. Clinical evidence on its efficacy remains limited, necessitating further research to validate its therapeutic benefits.
8 Surprising Reasons To Meditate 40 Minutes A Day That Can Change Your Life.pptx

8 Surprising Reasons To Meditate 40 Minutes A Day That Can Change Your Life.pptxHolistified Wellness
We’re talking about Vedic Meditation, a form of meditation that has been around for at least 5,000 years. Back then, the people who lived in the Indus Valley, now known as India and Pakistan, practised meditation as a fundamental part of daily life. This knowledge that has given us yoga and Ayurveda, was known as Veda, hence the name Vedic. And though there are some written records, the practice has been passed down verbally from generation to generation.TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kol...

TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kol...rightmanforbloodline
TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kolb, Ian Q. Whishaw, Verified Chapters 1 - 16, Complete Newest Versio
TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kolb, Ian Q. Whishaw, Verified Chapters 1 - 16, Complete Newest Version
TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kolb, Ian Q. Whishaw, Verified Chapters 1 - 16, Complete Newest VersionThe Best Ayurvedic Antacid Tablets in India

Treat the symptoms of indigestion, heartburn and stomach reflux with the 10 Best Ayurvedic Antacid Tablets in India.
A Classical Text Review on Basavarajeeyam

Basavarajeeyam is a Sreshta Sangraha grantha (Compiled book ), written by Neelkanta kotturu Basavaraja Virachita. It contains 25 Prakaranas, First 24 Chapters related to Rogas& 25th to Rasadravyas.
TEST BANK For Community Health Nursing A Canadian Perspective, 5th Edition by...

TEST BANK For Community Health Nursing A Canadian Perspective, 5th Edition by Stamler, Verified Chapters 1 - 33, Complete Newest Version Community Health Nursing A Canadian Perspective, 5th Edition by Stamler, Verified Chapters 1 - 33, Complete Newest Version Community Health Nursing A Canadian Perspective, 5th Edition by Stamler Community Health Nursing A Canadian Perspective, 5th Edition TEST BANK by Stamler Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Pdf Chapters Download Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Pdf Download Stuvia Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Study Guide Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Ebook Download Stuvia Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Questions and Answers Quizlet Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Studocu Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Quizlet Test Bank For Community Health Nursing A Canadian Perspective, 5th Edition Stuvia Community Health Nursing A Canadian Perspective, 5th Edition Pdf Chapters Download Community Health Nursing A Canadian Perspective, 5th Edition Pdf Download Course Hero Community Health Nursing A Canadian Perspective, 5th Edition Answers Quizlet Community Health Nursing A Canadian Perspective, 5th Edition Ebook Download Course hero Community Health Nursing A Canadian Perspective, 5th Edition Questions and Answers Community Health Nursing A Canadian Perspective, 5th Edition Studocu Community Health Nursing A Canadian Perspective, 5th Edition Quizlet Community Health Nursing A Canadian Perspective, 5th Edition Stuvia Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Pdf Chapters Download Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Pdf Download Stuvia Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Study Guide Questions and Answers Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Ebook Download Stuvia Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Questions Quizlet Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Studocu Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Quizlet Community Health Nursing A Canadian Perspective, 5th Edition Test Bank Stuvia
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Kat...

TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Kat...rightmanforbloodline
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Katzung, Verified Chapters 1 - 66, Complete Newest Version.
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Katzung, Verified Chapters 1 - 66, Complete Newest Version.
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Katzung, Verified Chapters 1 - 66, Complete Newest Version.
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Katzung, Verified Chapters 1 - 66, Complete Newest Version.REGULATION FOR COMBINATION PRODUCTS AND MEDICAL DEVICES.pptx

It includes regulation of combination products and medical devices. FDA and industry liaisons.
Histololgy of Female Reproductive System.pptx

Dive into an in-depth exploration of the histological structure of female reproductive system with this comprehensive lecture. Presented by Dr. Ayesha Irfan, Assistant Professor of Anatomy, this presentation covers the Gross anatomy and functional histology of the female reproductive organs. Ideal for students, educators, and anyone interested in medical science, this lecture provides clear explanations, detailed diagrams, and valuable insights into female reproductive system. Enhance your knowledge and understanding of this essential aspect of human biology.
Journal Article Review on Rasamanikya

Rasamanikya is a excellent preparation in the field of Rasashastra, it is used in various Kushtha Roga, Shwasa, Vicharchika, Bhagandara, Vatarakta, and Phiranga Roga. In this article Preparation& Comparative analytical profile for both Formulationon i.e Rasamanikya prepared by Kushmanda swarasa & Churnodhaka Shodita Haratala. The study aims to provide insights into the comparative efficacy and analytical aspects of these formulations for enhanced therapeutic outcomes.
Clinic ^%[+27633867063*Abortion Pills For Sale In Tembisa Central

Clinic ^%[+27633867063*Abortion Pills For Sale In Tembisa Central Clinic ^%[+27633867063*Abortion Pills For Sale In Tembisa CentralClinic ^%[+27633867063*Abortion Pills For Sale In Tembisa CentralClinic ^%[+27633867063*Abortion Pills For Sale In Tembisa CentralClinic ^%[+27633867063*Abortion Pills For Sale In Tembisa Central
Efficacy of Avartana Sneha in Ayurveda

Avartana Sneha is a unique method of Preparation of Sneha Kalpana in Ayurveda, mainly it is indicated for the Vataja rogas.
Cell Therapy Expansion and Challenges in Autoimmune Disease

There is increasing confidence that cell therapies will soon play a role in the treatment of autoimmune disorders, but the extent of this impact remains to be seen. Early readouts on autologous CAR-Ts in lupus are encouraging, but manufacturing and cost limitations are likely to restrict access to highly refractory patients. Allogeneic CAR-Ts have the potential to broaden access to earlier lines of treatment due to their inherent cost benefits, however they will need to demonstrate comparable or improved efficacy to established modalities.
In addition to infrastructure and capacity constraints, CAR-Ts face a very different risk-benefit dynamic in autoimmune compared to oncology, highlighting the need for tolerable therapies with low adverse event risk. CAR-NK and Treg-based therapies are also being developed in certain autoimmune disorders and may demonstrate favorable safety profiles. Several novel non-cell therapies such as bispecific antibodies, nanobodies, and RNAi drugs, may also offer future alternative competitive solutions with variable value propositions.
Widespread adoption of cell therapies will not only require strong efficacy and safety data, but also adapted pricing and access strategies. At oncology-based price points, CAR-Ts are unlikely to achieve broad market access in autoimmune disorders, with eligible patient populations that are potentially orders of magnitude greater than the number of currently addressable cancer patients. Developers have made strides towards reducing cell therapy COGS while improving manufacturing efficiency, but payors will inevitably restrict access until more sustainable pricing is achieved.
Despite these headwinds, industry leaders and investors remain confident that cell therapies are poised to address significant unmet need in patients suffering from autoimmune disorders. However, the extent of this impact on the treatment landscape remains to be seen, as the industry rapidly approaches an inflection point.
Recently uploaded (20)
Role of Mukta Pishti in the Management of Hyperthyroidism

Role of Mukta Pishti in the Management of Hyperthyroidism
8 Surprising Reasons To Meditate 40 Minutes A Day That Can Change Your Life.pptx

8 Surprising Reasons To Meditate 40 Minutes A Day That Can Change Your Life.pptx
Tests for analysis of different pharmaceutical.pptx

Tests for analysis of different pharmaceutical.pptx
TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kol...

TEST BANK For An Introduction to Brain and Behavior, 7th Edition by Bryan Kol...
TEST BANK For Community Health Nursing A Canadian Perspective, 5th Edition by...

TEST BANK For Community Health Nursing A Canadian Perspective, 5th Edition by...
TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Kat...

TEST BANK For Basic and Clinical Pharmacology, 14th Edition by Bertram G. Kat...
REGULATION FOR COMBINATION PRODUCTS AND MEDICAL DEVICES.pptx

REGULATION FOR COMBINATION PRODUCTS AND MEDICAL DEVICES.pptx
Muscles of Mastication by Dr. Rabia Inam Gandapore.pptx

Muscles of Mastication by Dr. Rabia Inam Gandapore.pptx
CHEMOTHERAPY_RDP_CHAPTER 6_Anti Malarial Drugs.pdf

CHEMOTHERAPY_RDP_CHAPTER 6_Anti Malarial Drugs.pdf
Clinic ^%[+27633867063*Abortion Pills For Sale In Tembisa Central

Clinic ^%[+27633867063*Abortion Pills For Sale In Tembisa Central
Cell Therapy Expansion and Challenges in Autoimmune Disease

Cell Therapy Expansion and Challenges in Autoimmune Disease
Fit for fitting: Bionext-DSS, a dataset of similars sites
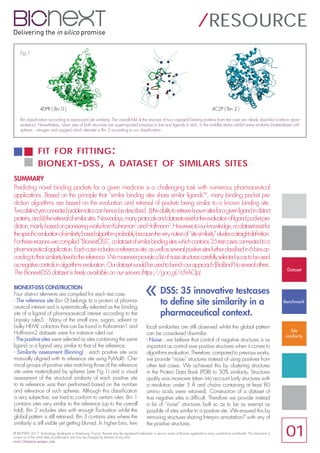
- 1. BIONEXT-DSS CONSTRUCTION Four distinct elements are compiled for each test case: - The reference site (bin 0) belongs to a protein of pharma- ceutical interest and is systematically selected as the binding site of a ligand of pharmaceutical interest according to the Linpisky rules5. Many of the small ions, sugars, solvent or bulky HEME cofactors that can be found in Kahraman1 and Hoffmann2 datasets were for instance ruled out. - The positive sites were selected as sites containing the same ligand or a ligand very similar to that of the reference. - Similarity assessment (Binning) : each positive site was manually aligned with its reference site using PyMol6. Che- mical groups of positive sites matching those of the reference site were materialized by spheres (see Fig.1) and a visual assessment of the structural similarity of each positive site to its reference was then performed based on the number and relevance of such spheres. Although this classification is very subjective, we tried to conform to certain rules: Bin 1 contains sites very similar to the reference (up to the overall fold); Bin 2 includes sites with enough fluctuation whilst the global pattern is still retained; Bin 3 contains sites where the similarity is still visible yet getting blurred. In higher bins, few local similarities are still observed whilst the global pattern can be considered dissimilar. - Noise : we believe that control of negative structures is as important as control over positive structures when it comes to algorithms evaluation. Therefore, compared to previous works, we provide “noise” structures instead of using positives from other test cases. We achieved this by clustering structures in the Protein Data Bank (PDB) to 30% similarity. Structures quality was moreover taken into account (only structures with a resolution under 3 Å and chains containing at least 80 amino acids were retained). Construction of a dataset of true negative sites is difficult. Therefore we provide instead a list of “noise” structures built so as to be as exempt as possible of sites similar to a positive site. We ensured this by removing structures sharing Interpro annotation7 with any of the positive structures. FIT FOR FITTING: BIONEXT-DSS, A DATASET OF SIMILARS SITES / RESOURCE SUMMARY Predicting novel binding pockets for a given medicine is a challenging task with numerous pharmaceutical applications. Based on the principle that “similar binding sites share similar ligands”4 , many binding pocket pre- diction algorithms are based on the evaluation and retrieval of pockets being similar to a known binding site. Twodistinctyetconnectedproblematicscanhencebedescribed:(i)theabilitytoretrieveknownsitesforagivenligandindistinct proteins,and(ii)theretrievalofsimilarsites.Nowadays,manyprotocolsanddatasetsexistfortheevaluationofligandpocketpre- diction,mainlybasedonpioneeringworksfromKahraman1 andHoffmann2 .However,toourknowledge,nodatasetexistfor thespecificevaluationofsimilaritybasedalgorithmprobablybecausetheverynotionof“sitesimilarity”eludesastraightdefinition. Forthesereasonswecompiled“Bionext-DSS”,adatasetofsimilarbindingsiteswhichcontains35testcasesconnectedtoa pharmaceuticalapplication.Eachcaseincludesareferencesite,aswellasseveralpositivesitesfurtherclassifiedin6binsac- cordingtotheirsimilarityleveltothereference.Wemoreoverprovidealistofnoisestructurescarefullyselectedsoastobeused asnegativecontrolsinalgorithmsevaluation.Ourdatasetwouldbeusedtobenchourapproach(BioBind3 )toseveralothers. The Bionext-DSS dataset is freely available on our servers (https://goo.gl/n5rAOp). Fig.1 DSS: 35 innovative testcases to define site similarity in a pharmaceutical context. « © BIONEXT 2017. Technology developed in Strasbourg, France. Bionext may be registered trademarks or service marks of Bionext registered in many jurisdictions worldwide. This document is current as of the initial date of publication and may be changed by Bionext at any time. HTTPS:/ /BIOSIGHT.BIO NEXT.COM 01 Dataset Benchmark Site similarity Bin classification according to eye-scored site similarity. The overall fold of the structure of two captopril binding proteins from test case are clearly dissimilar (cartoon repre- sentation). Nevertheless, when sites of both structures are superimposed (residues in line and ligands in stick, in the middle) atoms exhibit some similarity (materialized with spheres : nitrogen and oxygen) which denotes a Bin 2 according to our classification. 4DPR ( Bin 0 ) 4C2P ( Bin 2 )
- 2. DSS can be used for the two aforementioned problematics : - In order to evaluate the effectiveness of similarity retrieval, one can use the reference site as query and probe the ability to retrieve positive sites versus noise through the measurement of the area under the ROC curve (AUC). - In order to evaluate the algorithms ability to retrieve all known pockets of a given ligand, one can use alternatively all positive sites of a test case as a query and calculate an average AUC. As a complement, the structural alignment of positive sites with the reference provided in DSS can be used to dis- criminate valid prediction of positive sites. FACTS AND COMPARISONS For comparisons we applied our method for the evalua- tion of site similarity to the sites of the Kahraman and Hoffmann datasets. As can be seen in Table 1, DSS contains a larger amount of test cases as well as a larger amount of positive sites. Fig. 2 moreover reveals that DSS contains a bigger pro- portion of trivial similarities (bins 1 and 2), making our dataset more suited to evaluate algorithms on their ability to detect similarity. Moreover, the amount of difficult cases (bins 3 to 5) in DSS is comparable to that in Kahraman and Hoffmann, which makes our dataset also fit for the assessment of the more classical problematics of ligand binding pocket retrieval. With this dataset, we propose an experimental definition of similarity compliant to the intuition of a pharmaceuti- cally trained bioinformatician. To our knowledge, this is the first effort in that direction, and we believe this work is necessary in order to tune similarity-based algorithms. Although our classification in similarity bins is necessarily subjective, experiments conducted with our in-house al- gorithm BioBind as well as with ProBis8 show that lower bins were retrieved first, thus confirming an agreement between our intuitive definition of similarity and these algorithms. © BIONEXT 2017. Technology developed in Strasbourg, France. Bionext may be registered trademarks or service marks of Bionext registered in many jurisdictions worldwide. This document is current as of the initial date of publication and may be changed by Bionext at any time. HTTPS:/ /BIOSIGHT.BIO NEXT.COM ABOUT BIONEXT BIONEXTisaFrenchbioinformaticscompanyfounded in 2009 and dedicated to the better understanding and treatment of various diseases. As such, BIONEXT focuses on pharmaceutical problematics with a ma- jor interest in guiding the prediction of biochemical compounds for pharmaceuticals targets. BIONEXT aims to accelerate and improve the effectiveness of drug development and biological research by developing programs and software-based tools to tackle problematics of prime importance in the phar- maceutical field, such as drug profile assessment or suggestion of drug repurposing. Our first application BioBind is based on the now accepted principle that similar receptors bind similar ligands. W W W.BIO NEXT.COM POSSIBLE USES OF BIONEXT-DSS Dataset Positives Sites Test cases Bionext-DSS Kahraman Hoffman 222 64 97 35 7 10 Table 1: Number of test cases and positive sites for the studied datasets. Fig. 2: Percentage of positive sites per bin for each dataset. Numbers on the top of each bar refer to the numbers of positive sites. Fig.3: Application of DSS to BioBind and ProBis algorithms. Dataset Benchmark Site similarity REFERENCES 1. KAHRAMAN, A. ET AL. (2007). SHAPE VARIATIO N IN PROTEIN BINDIN G PO CKETS AND THEIR LIGANDS. JOURNAL OF MOLECULAR BIOLO GY, 368(1), 283–301. 2. HOFFMAN N, B. ET AL. (2010). A NEW PROTEIN BINDIN G PO CKET SIMILARITY MEASURE BASED O N C OMPARISO N OF CLOUDS OF ATOMS IN 3D: APPLICATIO N TO LIGAND PREDICTIO N. BMC BIOINFORMATICS, 11, 99. 3. BIOBIND: ALG ORITHM PATENTED BY BIO NEXT TO C OMPARE AND CLASSIFY PHYSIC O-CHEMICAL SURFACES. CF OTHER BIO NEXT C OMMUNICATIO NS FOR MORE INFORMATIO N. 4. KLABUNDE, T. (2007). CHEMO GEN OMIC APPROACHES TO DRUG DISCOVERY: SIMILAR RECEPTORS BIND SIMILAR LIGANDS. BR J PHARMAC OL, 152(1), 5–7 5. LIPINSKI, C. A (2004). LEAD- AND DRUG-LIKE C OMPOUNDS: THE RULE-OF-FIVE REVOLUTIO N. DRUG DISC OV TODAY, DEC(4), 337-41 6. DELAN O W. (2002). THE PYMOL MOLECULAR GRAPHICS SYSTEM. SAN CARLOS,CA,USA: DELAN O SCIENTIFIC LLC. HTTP: / /PYMOL.SOURCEFORGE.NET . 7. ROBERT D.FIN N. (2016) INTERPRO IN 2017 - BEYO ND PROTEIN FAMILY AND DOMAIN AN N OTATIO NS. NUCLEIC ACIDS RES (2017), 45 (D1), D190-D199. 8. KO N C, J. ET AL. (2015) PROBIS-CHARMMIN G: WEB INTERFACE FOR PREDICTIO N AND OPTIMIZATIO N OF LIGANDS IN PROTEIN BINDING SITES. J. CHEM. INF. MODEL., 55, 2308-2314. 02