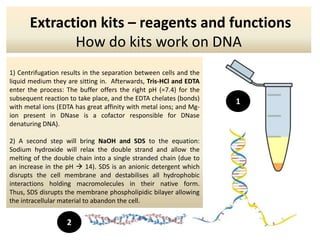
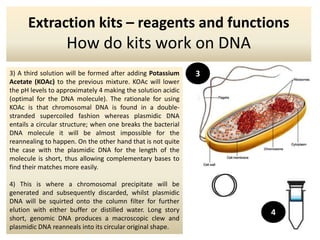
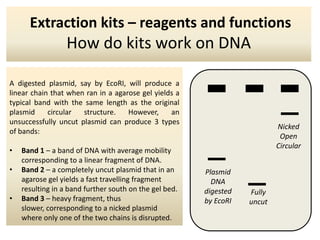
The document outlines the process of DNA extraction using various reagents and buffers, highlighting the roles of each component such as Tris-HCl, EDTA, NaOH, and SDS in separating plasmid DNA from chromosomal DNA. It describes the importance of maintaining optimal pH levels and the effects of different agents on DNA structure during extraction. Additionally, it details the formation of precipitates and how gel electrophoresis can be used to analyze the integrity of plasmid and chromosomal DNA post-extraction.