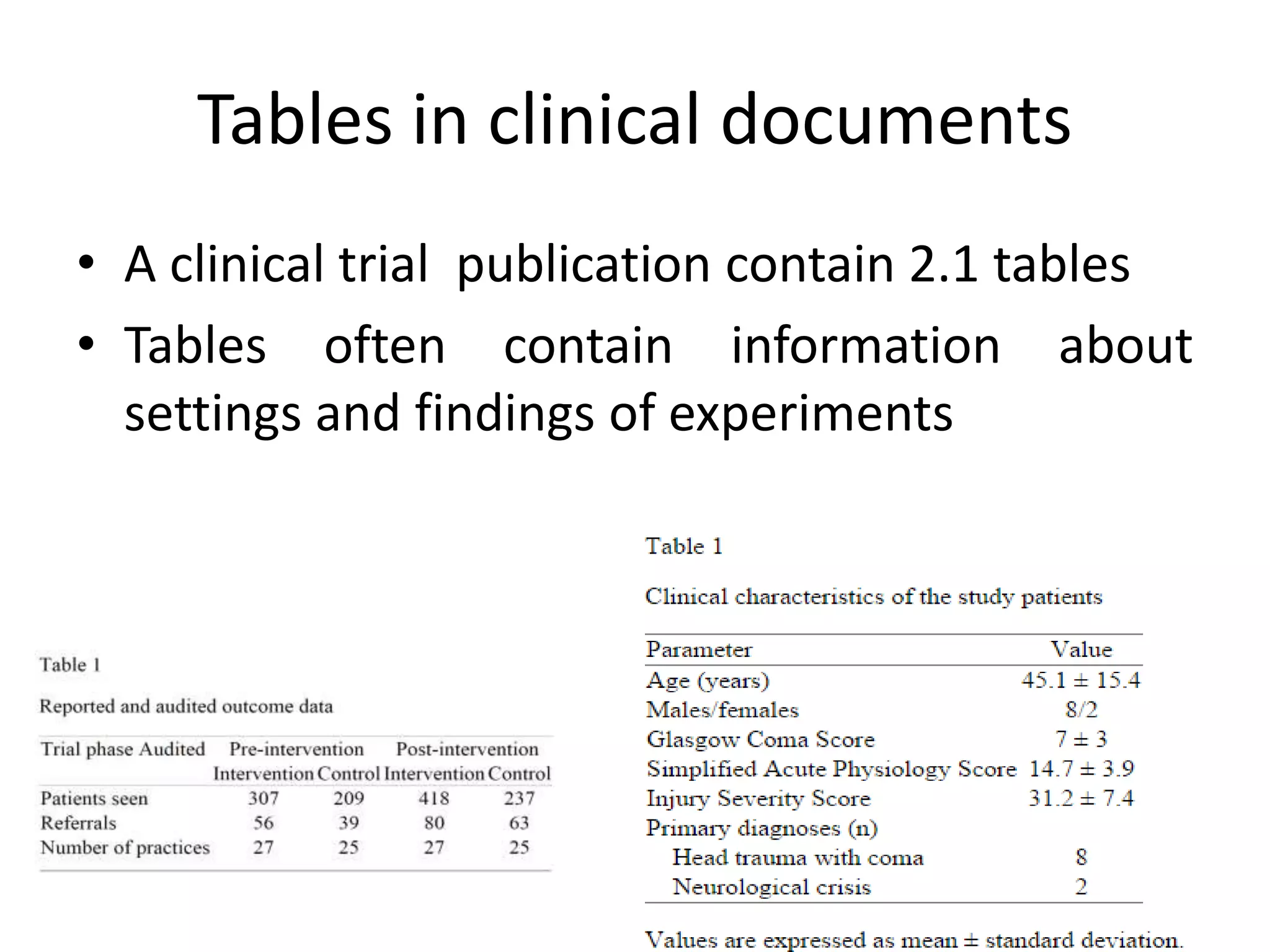
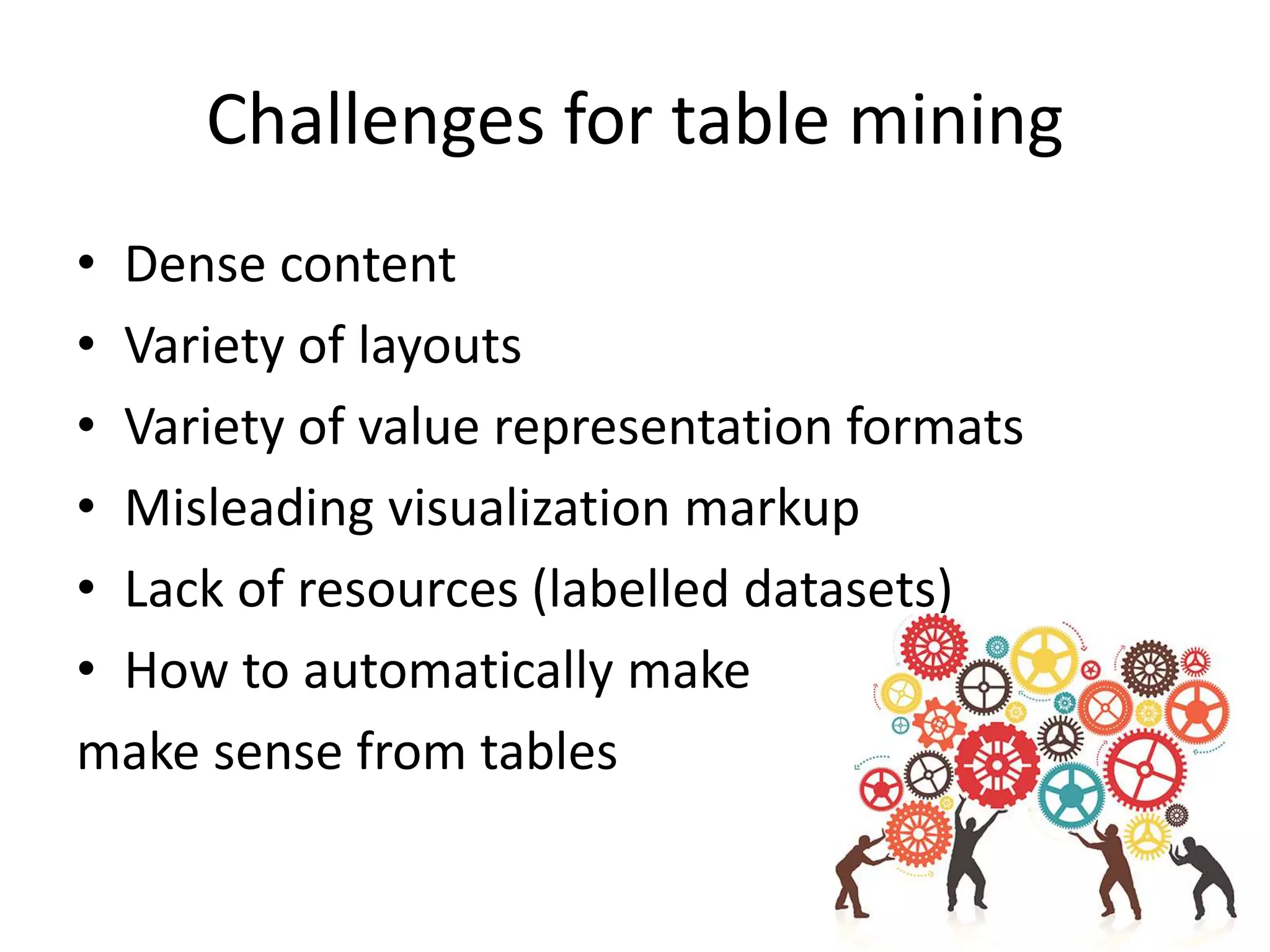
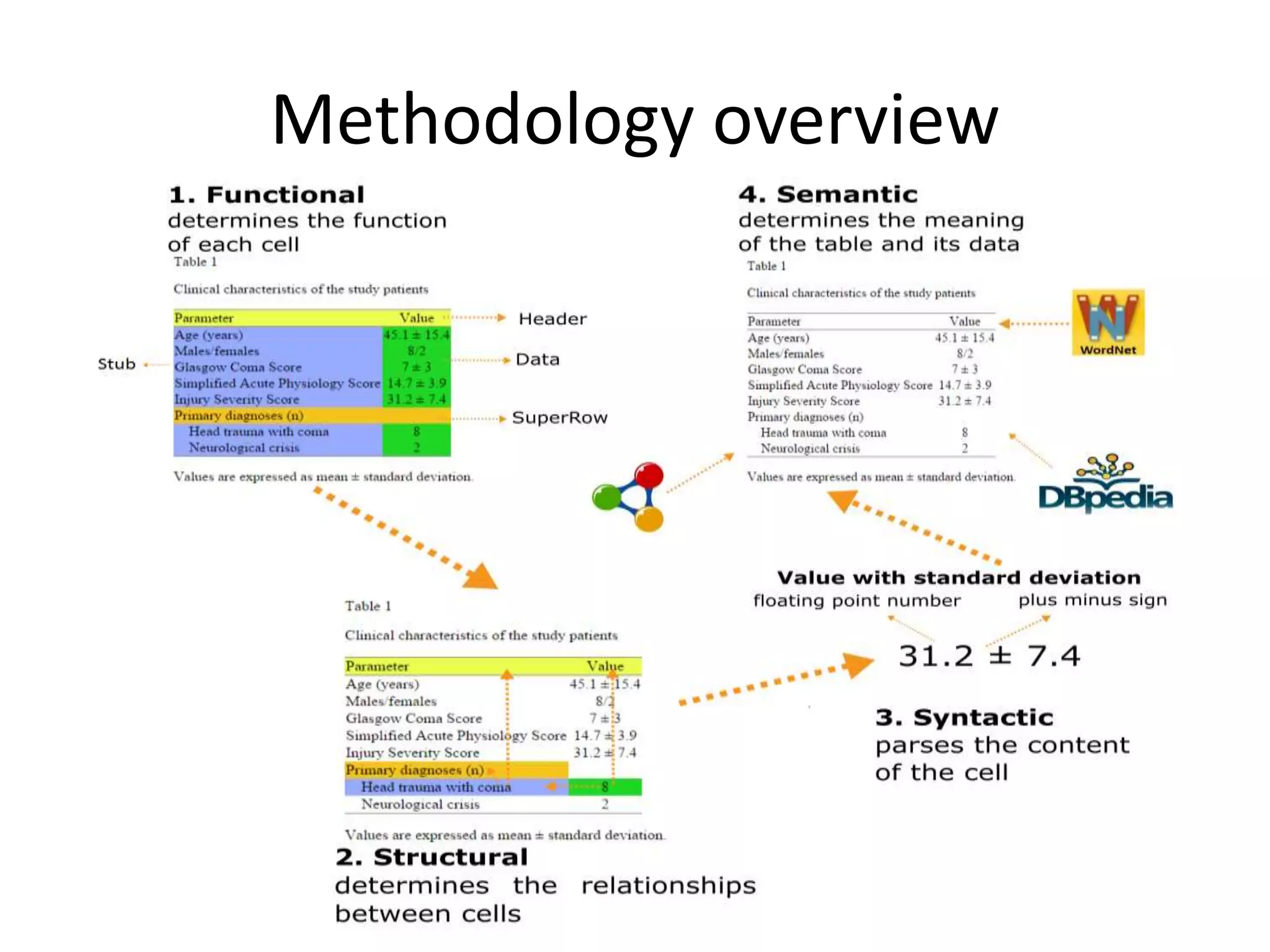
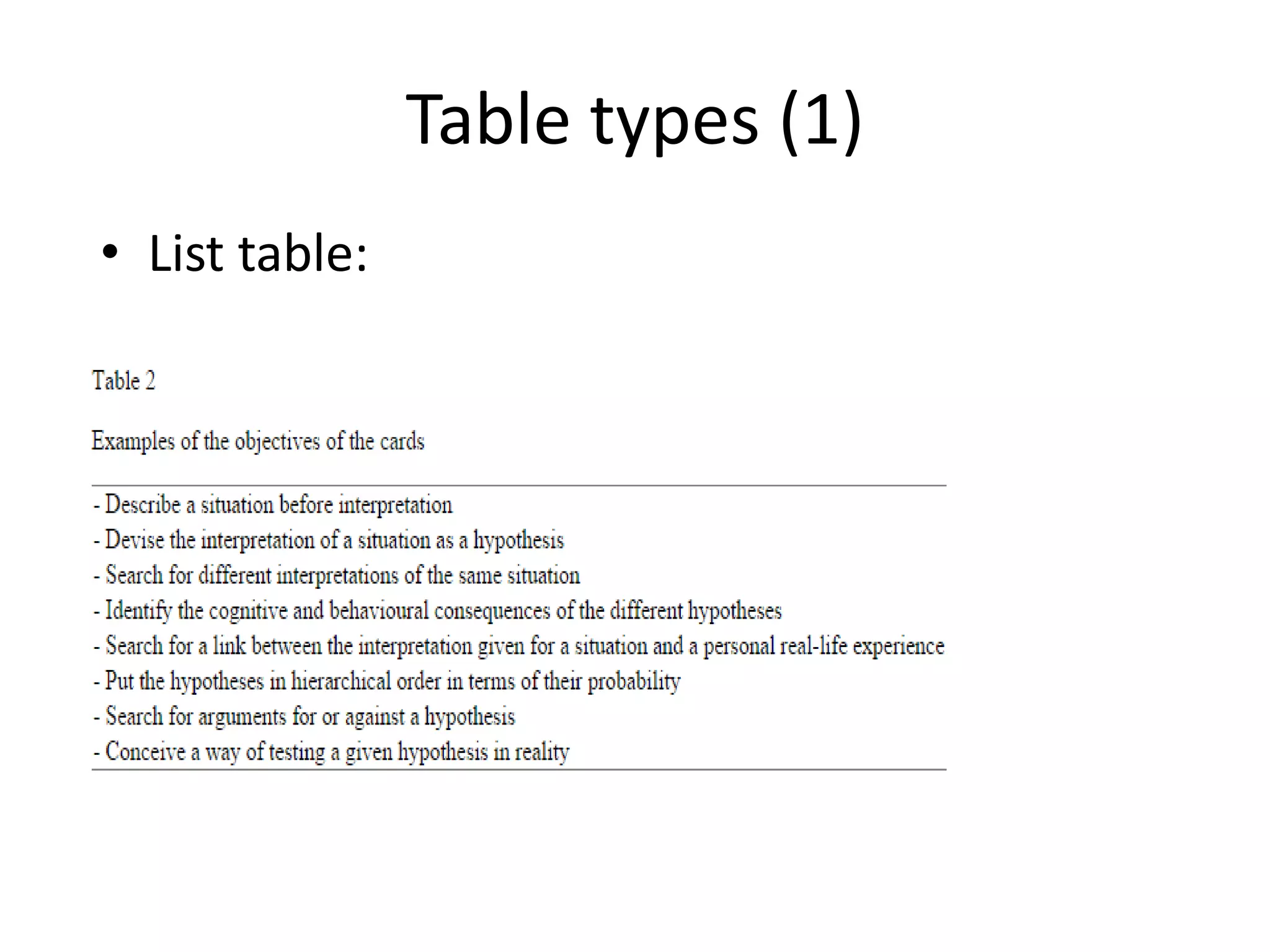
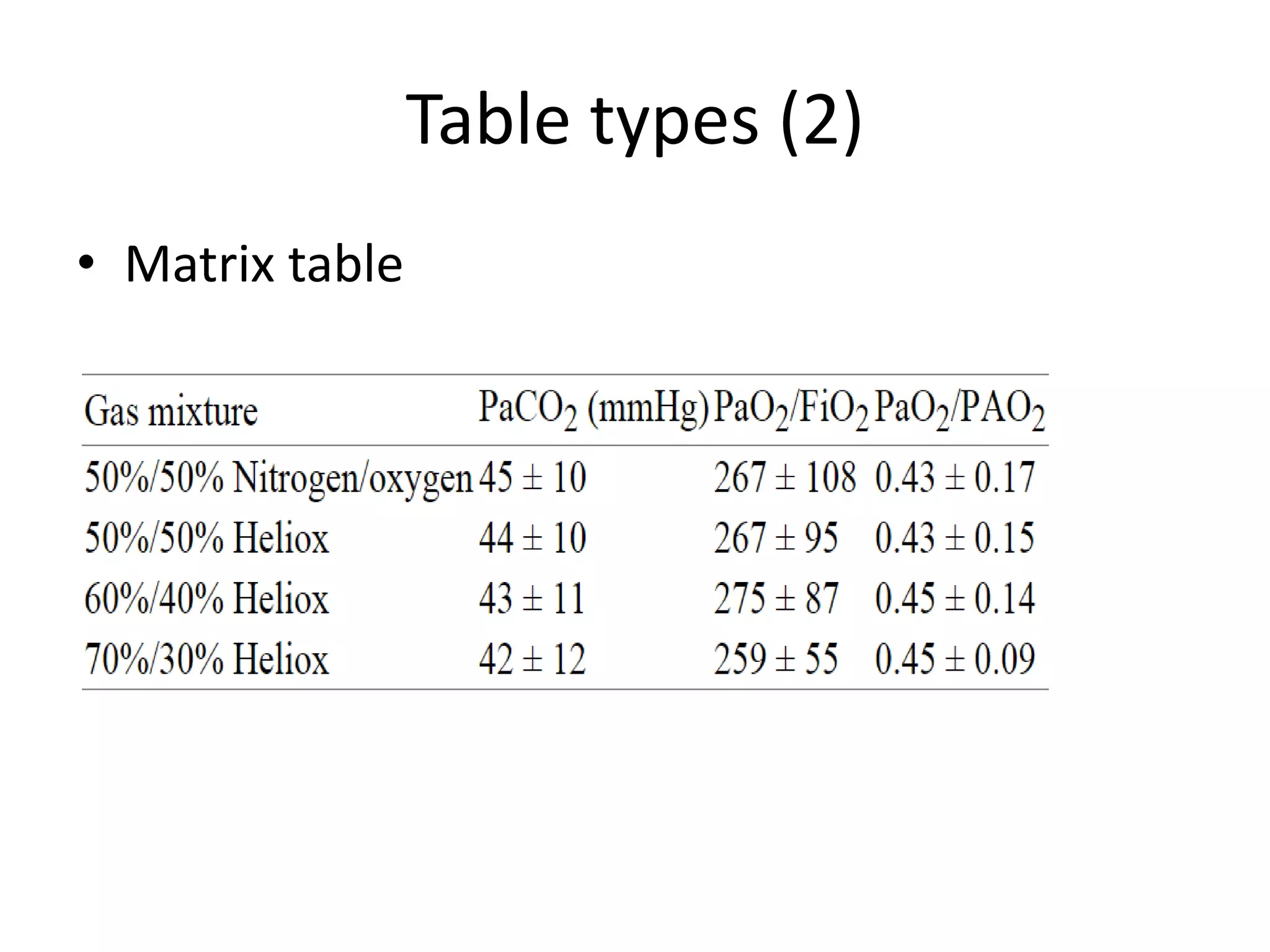
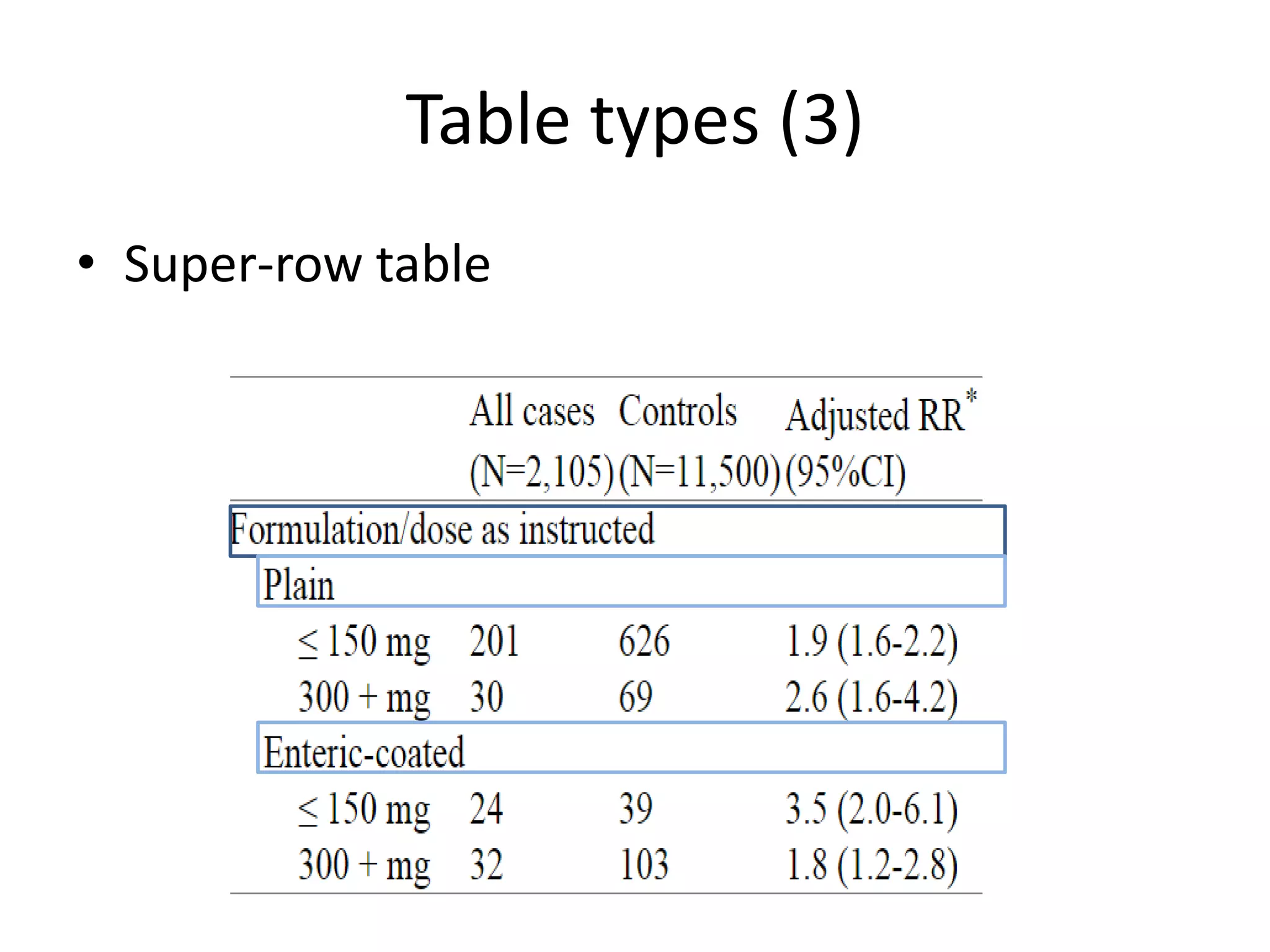
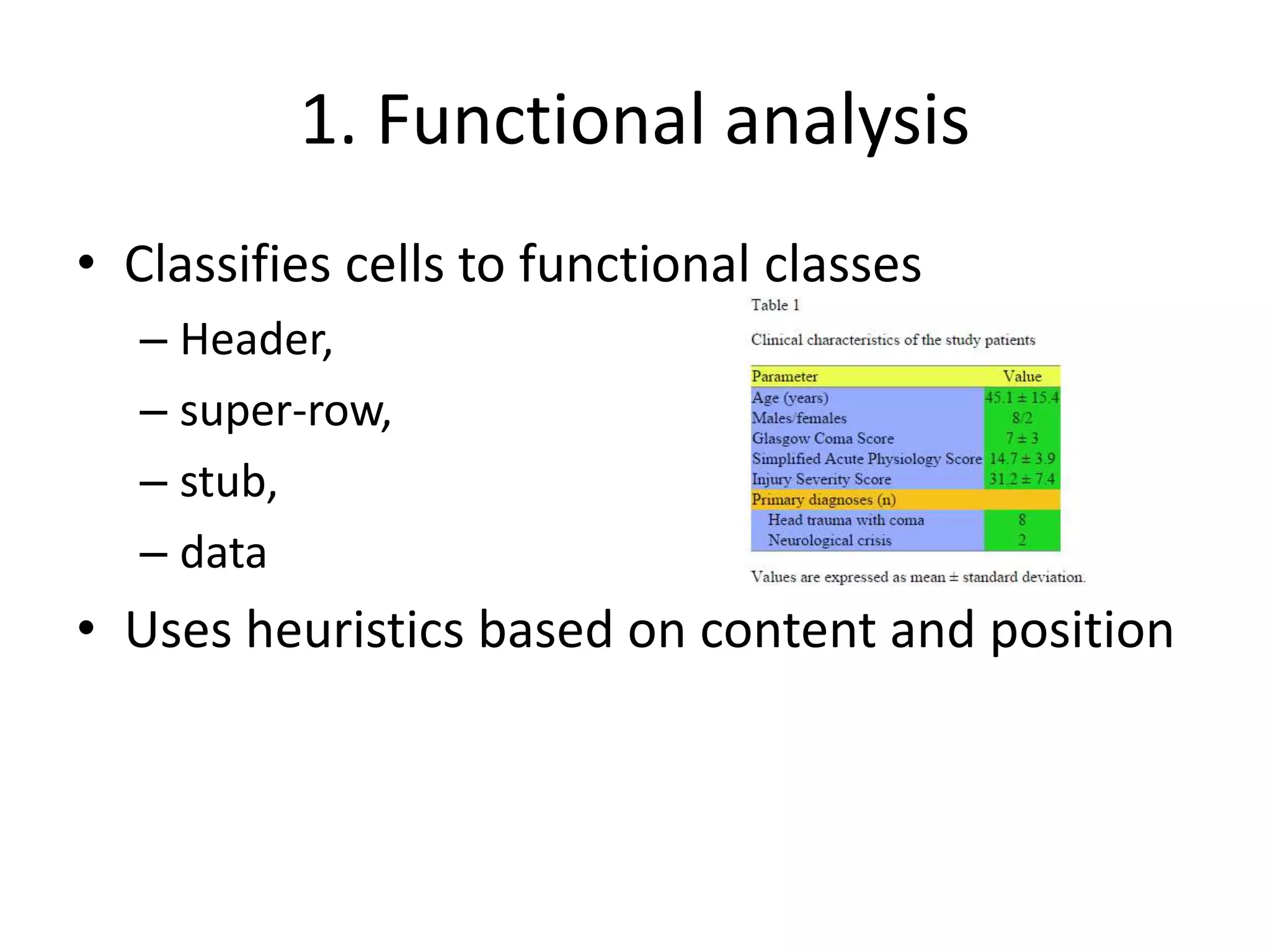
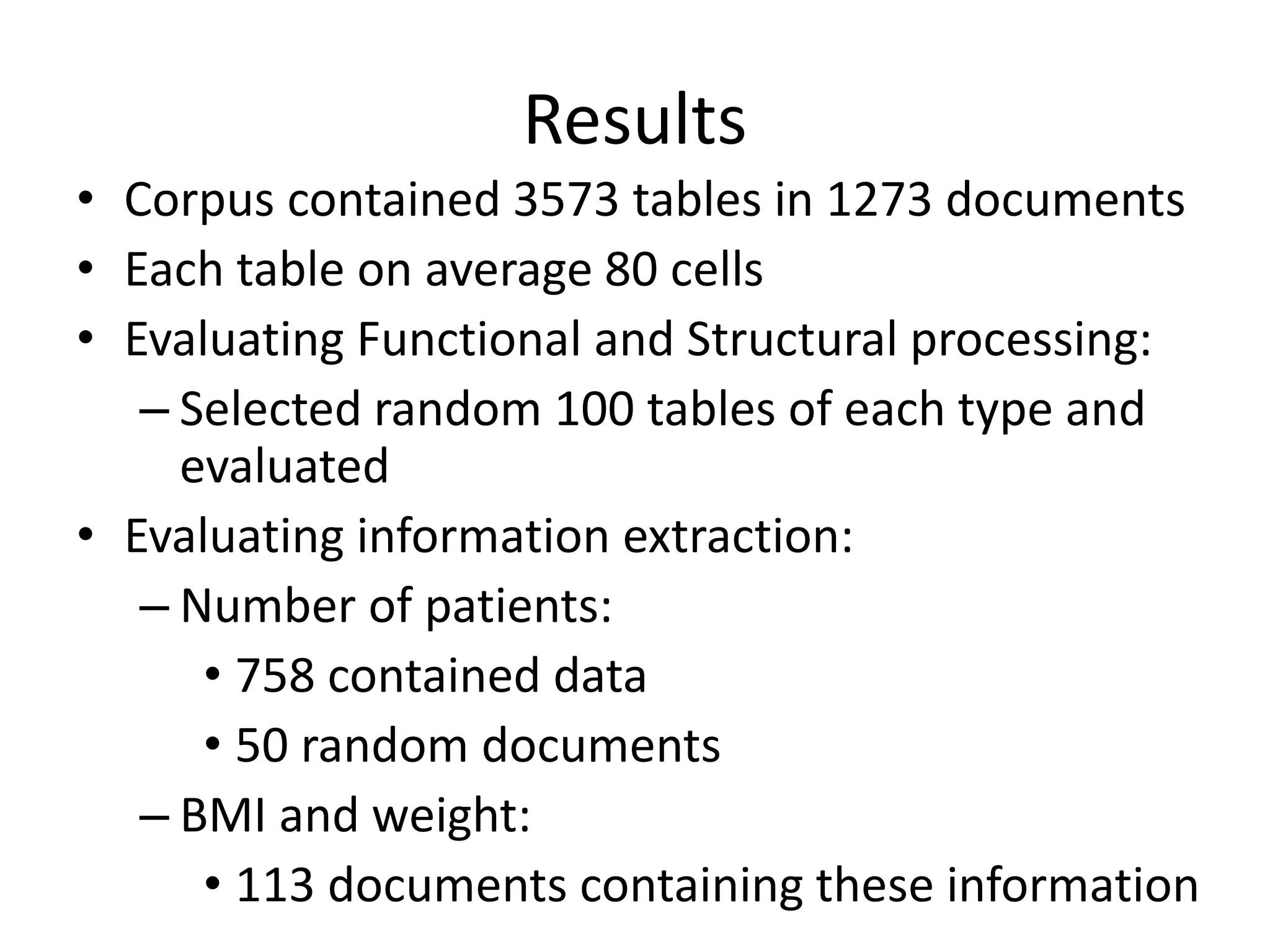
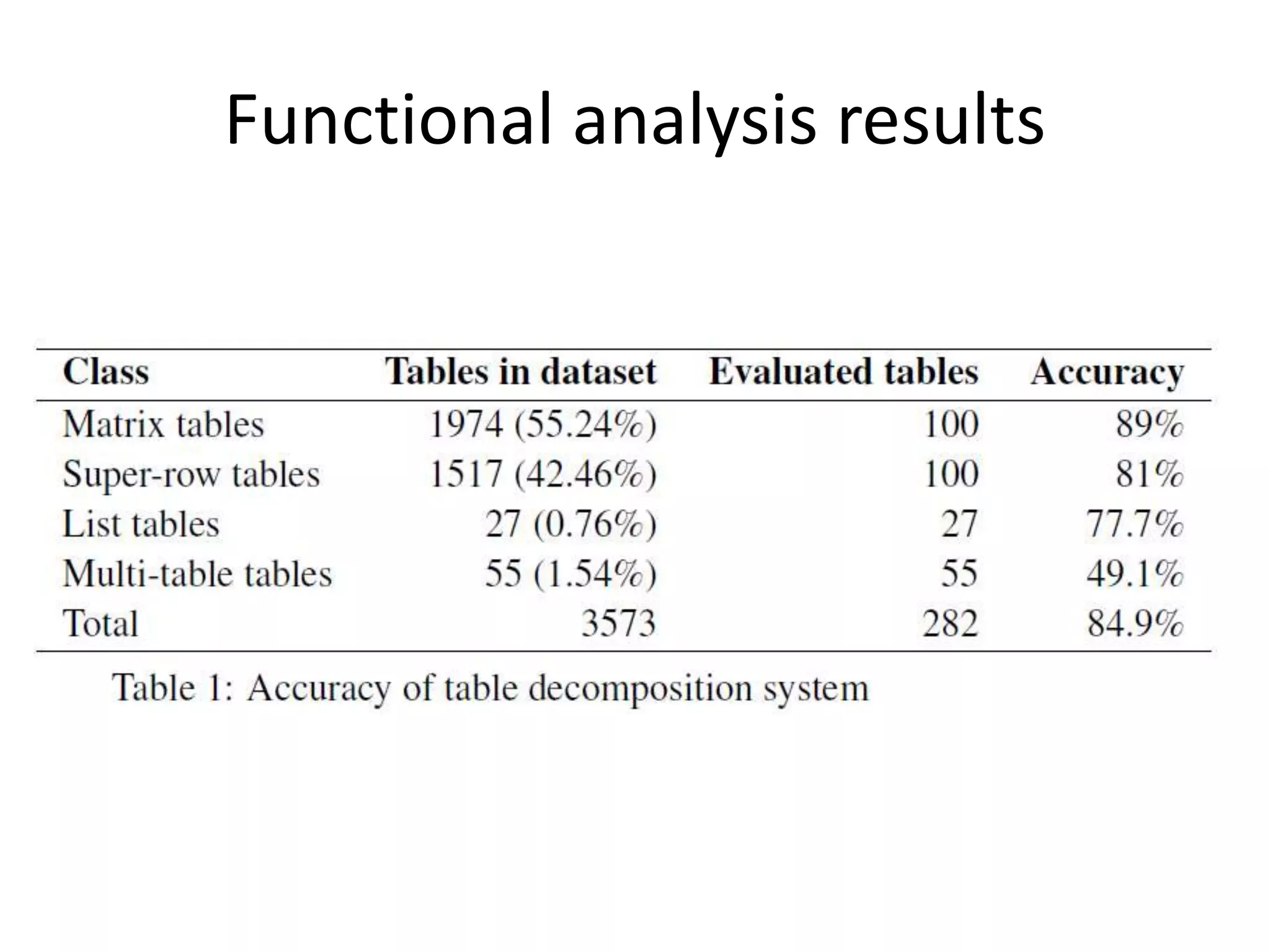
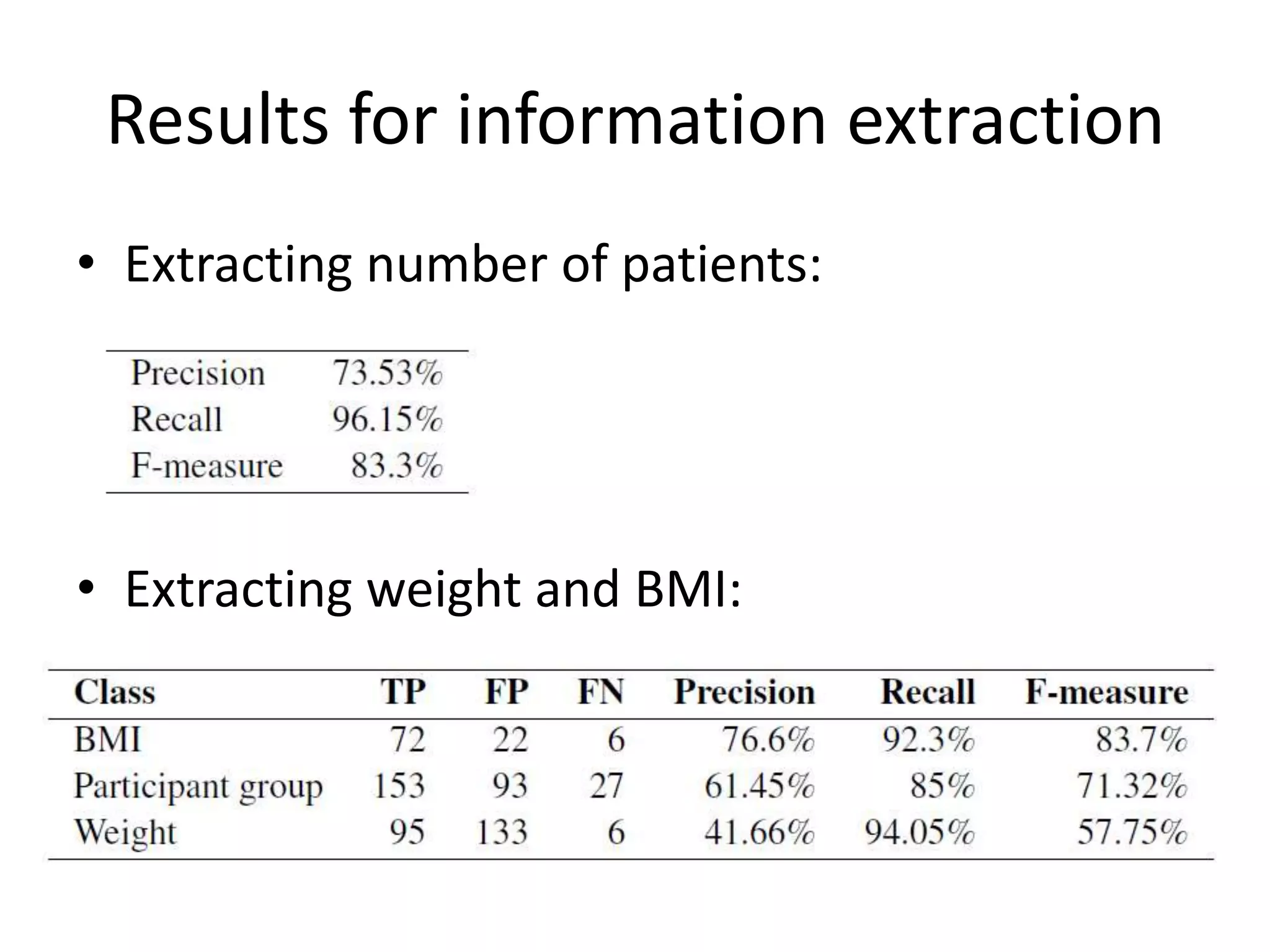
This document outlines a case study on the extraction of patient data, specifically BMI, weight, and patient numbers, from tables in clinical trial literature. It discusses the challenges of table mining, such as varied layouts and misleading visualizations, and presents a multi-layered rule-based methodology for information extraction. Results indicated promising performance in large-scale table mining, successfully classifying tables and extracting relevant data from 3,573 tables across 1,273 documents.