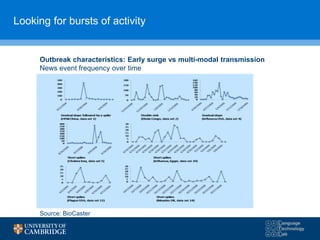
The document discusses the use of natural language processing (NLP) in digital disease informatics, highlighting its potential to convert unstructured health data into structured formats. It presents various case studies, including global infectious disease alerting and phenotype extraction, to demonstrate the effectiveness of NLP techniques in operational settings. Challenges such as data multilinguality and real-time scaling are also addressed, with an emphasis on the importance of robust data collection from diverse health-related text sources.






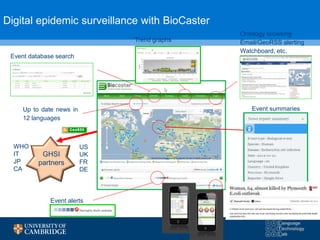
![BIOCASTER: GLOBAL INFECTIOUS DISEASE
ALERTING AND MAPPING
Case study #1
[5] Collier, N. et al. (2008). BioCaster: detecting public health rumors with a Web-based text mining system. Bioinformatics, 24(24), 2940-2941.
[6] Collier, N., et al. (2011). OMG U got flu? Analysis of shared health messages for bio-surveillance. J. Biomedical Semantics, 2(S-5), S9.
[7] Hay, S. I., et al. (2013). Global mapping of infectious disease. Philosophical Transactions of the Royal Society of London B: Biological
Sciences, 368(1614), 20120250.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-8-320.jpg)



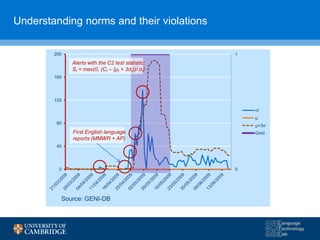
![5 detection algorithms
1. Early aberration reporting system (EARS) C2 algorithm
• captures the number of standard deviations that the current count exceeds the history mean;
• St = max(0, (Ct – (μt + kσt))/ σt)
2. EARS C3 algorithm
• similar to C2 except that C3 uses a weighted sum of the previous 3 days for the current period;
3. W2 algorithm
• a modified version of C2 which ignores history counts on Saturdays and Sundays to compensate for day of week effects;
4. F statistic
• compares the variance in the history window to the variance in the current window;
• St = σt
2 +σb
2
5. Exponential Weighted Moving Average (EWMA)
• provides less weight to days in the history that are further from the test day.
• St = (Yt – μt)/[σt * (λ/(2- λ))1/2], where Y1 = C1 and Yt = λCt + (1- λ)Yt-1
Model parameters were estimated based on an additional 5 epidemic data sets from ProMED-mail (data not
shown)
[8] Burkom H. S. (2005), “Accessible Alerting Algorithms for Biosurveillance”. National Syndromic Surveillance Conference
[9] Jackson M. L. et all (2007), “A simulation study comparing aberration detection algorithms for syndromic surveillance” Medical Informatics and Decision
Making , 7(6): BMC, DOI: 10.1186/1472-6947-7-6.
[10] Madoff L. (2004), “ProMED-mail: An early warning system for emerging diseases”. Clin Infect Dis , 39(2): 227–232.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-18-320.jpg)
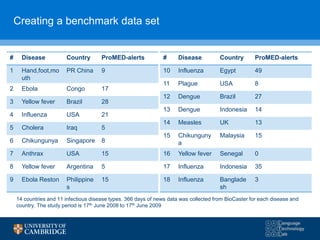
![C3 C2 W2 F-statistic EWMA
Sensitivity 0.74 0.66 0.66 0.78 0.73
(0.69-0.78) (0.61-0.72) (0.60-0.71) (0.74-0.82) (0.68-0.78)
Specificity 0.96 0.98 0.98 0.92 0.95
(0.95-0.96) (0.98-0.98) (0.98-0.99) (0.91-0.92) (0.94-0.96)
PPV 0.55 0.64 0.65 0.46 0.47
(0.98-0.99) (0.98-0.99) (0.98-0.99) (0.98-0.99) (0.98-0.99)
NPV 0.98 0.98 0.98 0.98 0.98
(0.98-0.99) (0.98-0.99) (0.98-0.99) (0.98-0.98) (0.98-0.99)
Alarms/100 days 6.48 4.52 4.17 12.34 7.85
F-measure 0.63 0.65 0.66 0.58 0.58
Results in parentheses show 95% confidence intervals
[11] Collier, N. (2009), “What’s unusual in online disease outbreak news?”, in BMC Biiomedical Semantics, 1(2).
Comparison of 5 aberration detection algorithms](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-20-320.jpg)
![Field evaluation
• (2006-2012) Global Health Security Initiative– a unique initiative by G7+WHO+EC to
bring together end-users, system providers and stakeholders to test the feasibility of
open source public health intelligence systems.
[12] Barboza, P., Vaillant, L., Le Strat, Y., Hartley, D. M., Nelson, N. P., Mawudeku, A., Madoff, L. C., Linge, J. P., Collier, N., Brownstein, J. S. and Astagneau,
P. (2014). Factors Influencing Performance of Internet-Based Biosurveillance Systems Used in Epidemic Intelligence for Early Detection of Infectious Diseases
Outbreaks. PloS one, 9(3), e90536.
[13] Barboza, P., Vaillant, L., Mawudeku, A., Nelson, N., Hartley, D., Madoff, L., Linge, J., Collier, N., Brownstein, J., Yangarber, R. and Astagneau, P. (2013),
“Evaluation of epidemic intelligence systems integrated in the Early Alerting and Reporting project for the detection of A/H5N1 Influenza events”, PLoS One,
8(3):e57252.
Major findings for A/H5H1:
- Detection rates for individual systems from
31% to 38%
- Rising to 72% for the combined system
- PPV ranged from 3% to 24%
- F1 ranged from 6% to 27%
- Sensitivity ranged from 38% to 72%
- Average improvement in alerting over WHO or
OIE was 10.2 days](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-21-320.jpg)

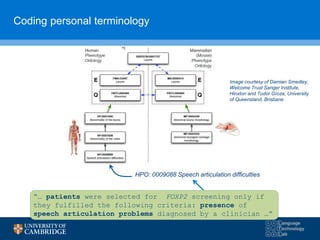
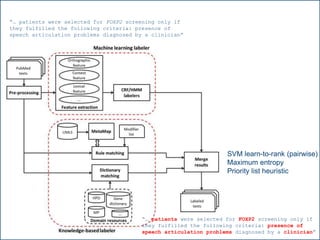
![PHENOMINER/PHENEBANK: EXTRACTING A
DATABASE OF PHENOTYPE TERMS
Case study #2
[14] Collier, N., Groza, T., Smedley, D., Robinson, P., Oellrich, A. and Rebholz-Schuhmann, D. (2015). PhenoMiner: from text to a database of
phenotypes associated with OMIM diseases. Database, Oxford University Press (in press).
[15] Collier, N., Oellrich, A. and Groza, T. (2013), “Toward knowledge support for analysis and interpretation of complex traits”, Genome Biology
14(9):214.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-23-320.jpg)
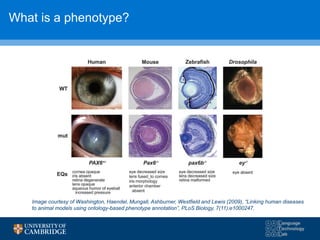
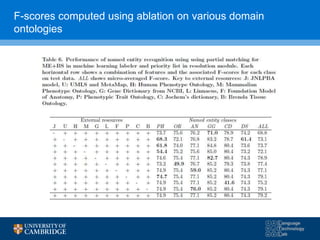
![F-scores using 3 hypothesis resolution strategies
[16] Collier, N., Tran, M., Le, H. Ha, Q., Oellrich, A. Rebholz-Schuhmann, D. (2013), “Learning to recognize phenotype candidates in the auto-immune literature
using SVM re-ranking”, PLoS One 8(10): e72965.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-29-320.jpg)
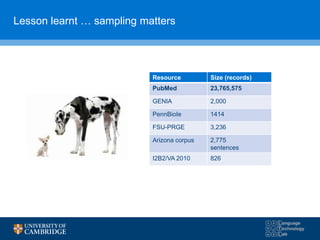
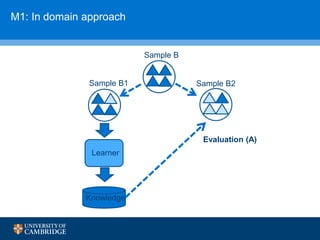
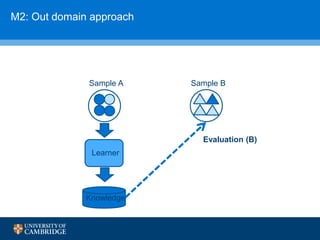
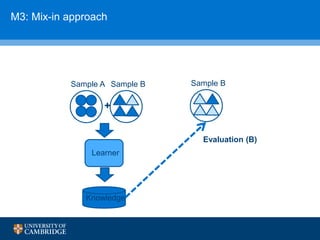
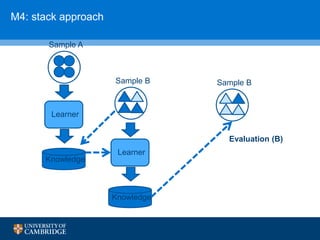
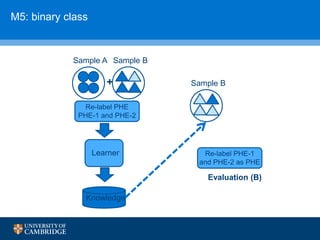
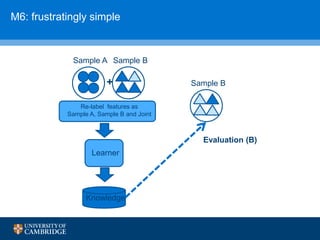
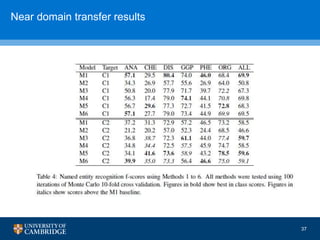
![How can we do domain adaptation better (with less
annotations)?
[17] Collier, N., Paster, F., Campus, H., & Tran, A. M. V. (2014), “The impact of near domain transfer on biomedical named entity recognition”, Proc. 5th
International Workshop on Health Text Mining and Information Analysis (LOUHI) at the European Conference on Computational Linguistics (EACL),
Gothenburg, Sweden, pp. 11-20.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-38-320.jpg)
![SIPHS: UNDERSTANDING THE VOICE OF THE
PATIENT
Case study #3
[18] Limsopatham, N. and Collier, N. (2015), “Adapting phrase-based machine translation to normalise medical terms in social media messages”,
in Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17-21 September 2015, pp.
1675-1680.
[19] Limsopatham, N. and Collier, N. (2015), “Towards the semantic interpretation of personal health messages from social media”, in Proceedings
of the 24th ACM International Conference on Information and Knowledge Management (CIKM 2015), Workshop on Understanding the City with
Urban Informatics (UCUI 2015), Melbourne, Australia, 19-23 October 2015.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-39-320.jpg)

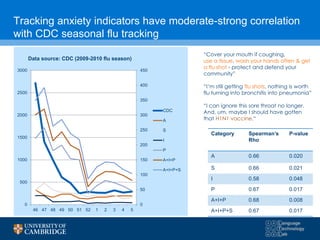
![Frustratingly simple models work better
Classifying respiratory syndrome: Turning 225,000 Tweets into a
high correlation influenza tracker
[22] Doan, S., Ohno-Machado, L. and Collier, N. (2012), "Enhancing Twitter data analysis with simple semantic filtering: example in tracking Influenza-Like Illnesses", in
the 2nd IEEE Conference on Healthcare Informatics, Imaging and Systems Biology: Analyzing Big Data for Healthcare and Biomedical Sciences, California, USA,
September 27-28.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-42-320.jpg)
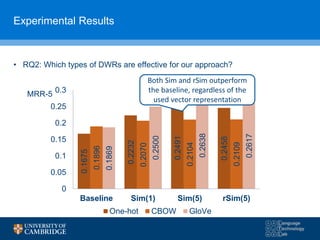
![“You shall know a word by the company it keeps”
– (Firth, J. R. 1957)
• Existing work [1,2] used word vector similarity to measure the
semantic similarity between texts
Performance seems depended on the used vector representation (e.g.
CBOW [1], GloVe [2])
[23] Mikolov et al. Distributed representations of words and phrases and their compositionality. NIPS 2013
[24] Pennington et al. GloVe: Global vectors for word representation. EMNLP 2014
• Recent advances in deep learning
technology [1,2] allowed the learned
representation of terms (i.e. DWRs) that
could capture the semantic similarity of
terms based on their co-occurrences e.g.
Continuous bag-of-words (CBOW) [1], Global
Vector (GloVe) [2]
44](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-44-320.jpg)
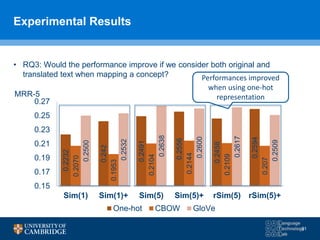
![Related work – Phrase-based MT
• Phrase-based MT [3]: Translate between languages by learning local
term dependencies from parallel corpora
We adapt phrase-based MT to translate from social media language to
formal medical language
Can’t even focus forreal no concentrate ???
[25] Koehn et al. Statistical phrase-based translation. NAACL 2003
45](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-45-320.jpg)
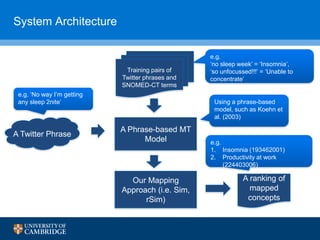
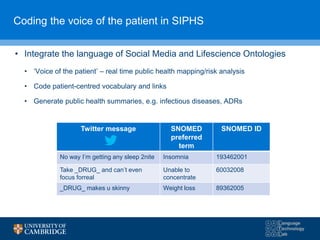
![Adapting Phrase-based MT for Twitter Normalisation
• We use phrase-based MT to translate social media text to formal
medical text, then map the translated symptoms to a SNOMED-CT
concept
Can’t even focus forreal unable to focus unable to concentrate
(ID 60032008)
translate
find semantic distance
[18] Limsopatham, N. and Collier, N. (2015), “Adapting phrase-based machine translation to noramlise medical terms
in social media messages”, in Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing,
Lisbon, Portugal, September, pp. 1675-1680.](https://image.slidesharecdn.com/warwick10-151019073907-lva1-app6891/85/Exploiting-NLP-for-Digital-Disease-Informatics-46-320.jpg)