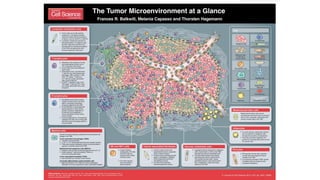
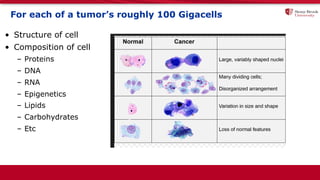
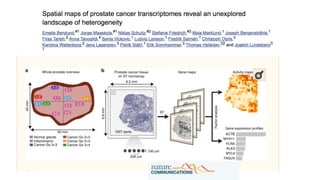
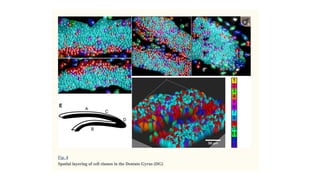
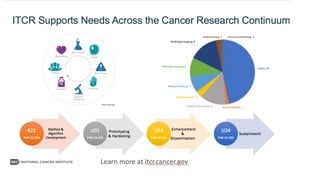
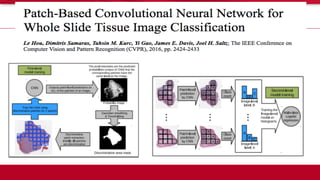
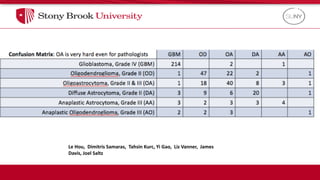
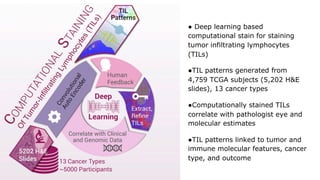
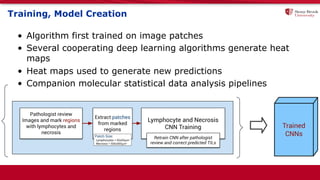
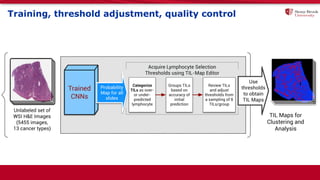
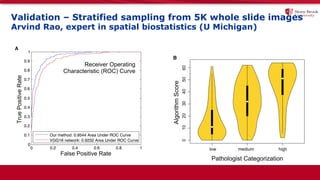
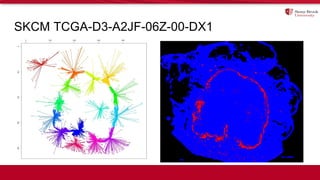
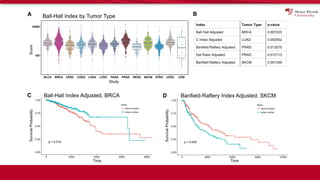
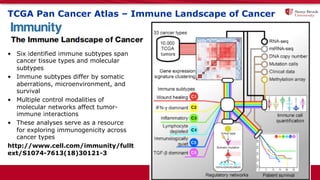
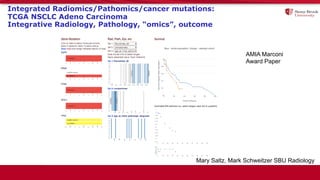
The document discusses the integration of deep learning and streaming data in cancer research, emphasizing the need for comprehensive analysis of tumor characteristics through digital pathology and imaging. It provides statistics on global cancer incidence and mortality, highlighting the role of various research networks and consortia in advancing cancer genomics and imaging. The future of digital pathology involves sophisticated algorithms that enhance image analysis and link molecular features to clinical outcomes.